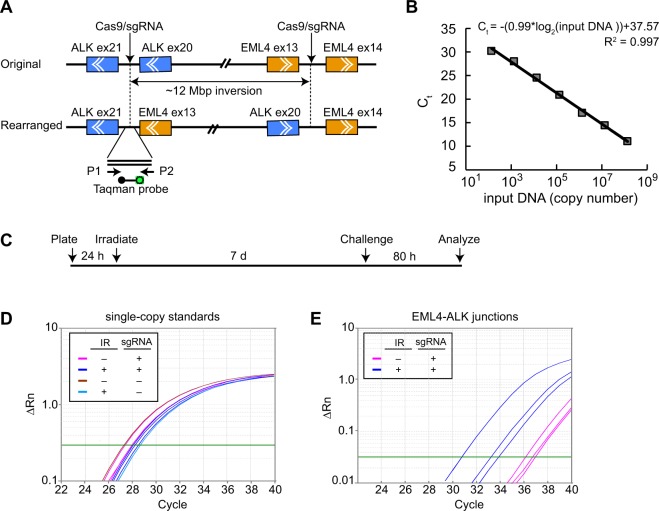
Figure 1.
Experimental strategy and example data. (A) Schematic depicting sites of Cas9/sgRNA cleavage relative to ALK and EML4 genes. Chevrons depict direction of transcription. Cleavage sites are within introns. Exon (ex) numbers are shown. P1 and P2 are primers for Taqman PCR assays. Taqman hybridization probe is shown with fluorophore (green) and quencher (black) that separate upon hybridization to amplicon. (B) Reconstruction experiment showing linearity of Taqman qPCR assay. (C) Experimental timeline showing irradiation, recovery, Cas9/sgRNA challenge, and harvesting of DNA for analysis. (D,E) Example Taqman PCR data. Independent cultures were irradiated, challenged, and DNA collected for analysis. Amplification curves are shown for single-copy internal standard (RNase P, Panel D) or for EML4-ALK junctions (Panel E). Irradiation was performed at an LET of 108 keV/μm). Parallel cultures not challenged with Cas9/sgRNAs were included in the experiment in Panel E but showed no detectable amplification products after 40 cycles of amplification. Plots depict normalized fluorescence reporter values (ΔRn) as a function of cycle number. Green line denotes software-determined threshold for determination of amplification (Ct) values.