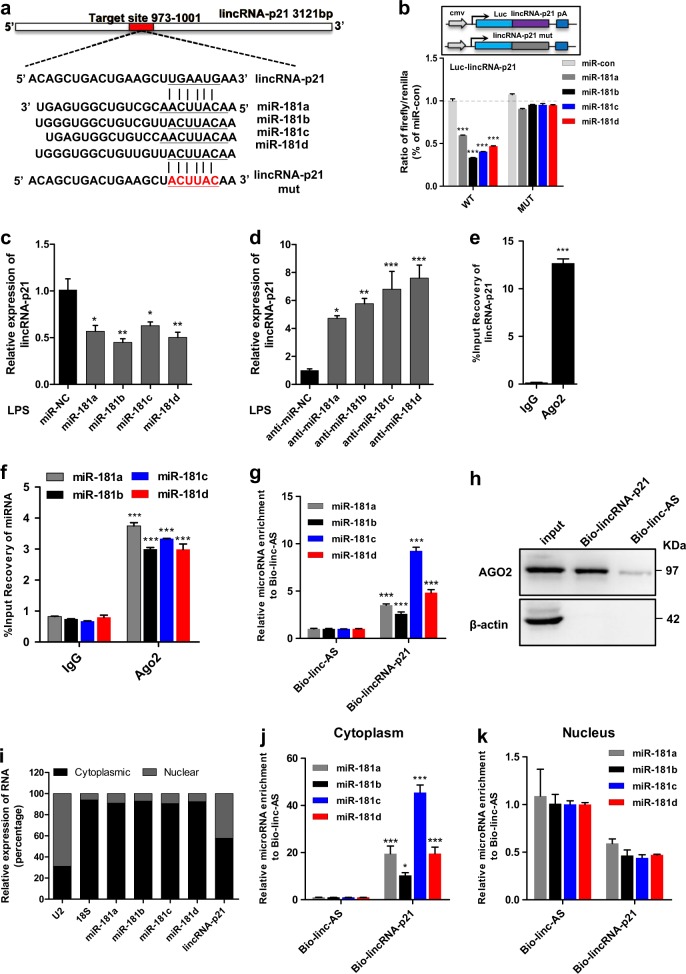
Fig. 3. LincRNA-p21 physically associates with the miR-181 family.
a Putative miR-181 family binding site in lincRNA-p21. The potential complementary residues are underlined and mutated bases are indicated in red. b Upper panel, schematic representation of pmirGLO-based luciferase reporter plasmid containing wild-type lincRNA-p21 (Luc-lincRNA-p21-wt) and a mutant reporter construct (Luc-lincRNA-p21-mut). Lower panel, relative luciferase activity of the luciferase constructs of Luc-lincRNA-p21-wt or Luc-lincRNA-p21-mut co-transfected with miR-control/miR-181 family in HEK293 cells. A representative experiment with triplicates of two independent experiments is shown. Data are expressed as mean ± SEM of triplicate wells. *P < 0.05, **P < 0.01 in one-way analysis of variance ANOVA followed by Bonferroni test. c, d qRT-PCR analysis of lincRNA-p21 in BV2 microglia cells treated with LPS following transfection of cells with miR-control/miR-181a, b, c, d mimics (c); or anti-miR-control/anti-miR-181 a, b, c, d (inhibitors) (d). Data are expressed as mean ± SEM (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 versus control using one-way analysis of variance ANOVA followed by Bonferroni test. e, f Anti-Ago2 RIP was performed in BV2 microglia cell lysates. Co-precipitated RNAs were used to detect the relative enrichment of lincRNA-p21 (e) and miR-181 family (f) associated with Ago2, compared with that in normal IgG control. g, h In vitro-synthesized biotin-labeled RNA pulldown experiments in BV2 microglia cells. BV2 microglia cell lysates were incubated with in vitro-synthesized biotin-labeled lincRNA-p21 sense or antisense RNA for biotin pull-down assay, followed by qRT-PCR to detect miR-181 family (g) and western blotting to detect Ago2 (h) associated with lincRNA-p21. Bio-linc-AS, biotinylated-lincRNA-p21-antisense RNA; bio-lincRNA-p21, biotinylated-lincRNA-p21-sense RNA. i Representative analysis of lincRNA-p21 and miR-181 family distribution in nuclear and cytoplasmic fractions in BV2 microglia cells. U2 snRNA and 18S rRNA served as controls for nuclear and cytoplasmic RNA, respectively. Exogenous spike-in of cel-mir-39 served as a normalizer for the qRT-PCR analysis. j, k Biotin pull-down assay with biotin-labeled lincRNA-p21 sense or antisense RNA was performed with the cellular fractions of cytoplasm (j) or nucleus (k) of BV2 microglia cells. A representative experiment with triplicates of two independent experiments is shown. Data are expressed as mean ± SEM of triplicate wells. *P < 0.05, **P < 0.01 ***P < 0.001 in two-way analysis of variance ANOVA followed by Bonferroni test