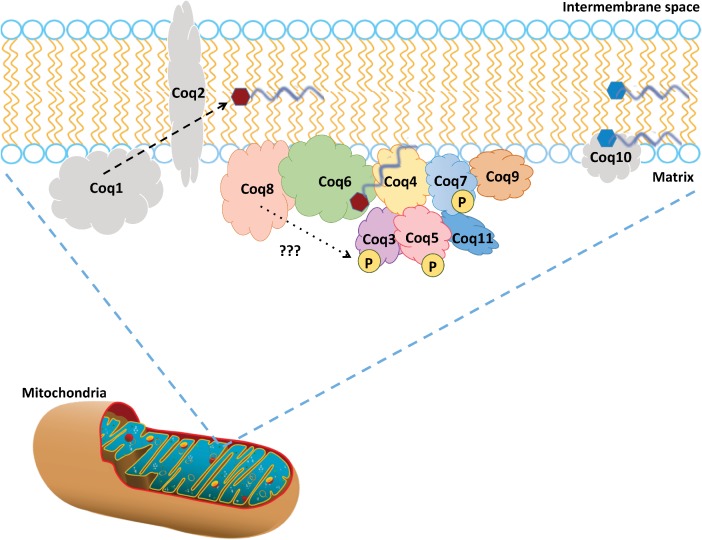
Figure 2. A model of the CoQ Synthome in the yeast S. cerevisiae.
Studies in S. cerevisiae have provided evidence for a high-molecular mass multisubunit protein and lipid complex, the CoQ synthome (see text for references). The Coq3–Coq9 and Coq11 polypeptides, designated in color, co-purify, and are members of this complex that is peripherally associated with the matrix-side of the inner mitochondrial membrane. Coq1, Coq2, and Coq10 are individual polypeptides that do not associate with the complex (indicated in gray). Coq1 and Coq2 synthesize the early intermediates HHB and HAB (denoted by red hexagon with a gray hexaprenyl tail). Coq10 binds CoQ (and also late-stage CoQ-intermediates denoted as blue hexagons with a gray tail), and functions as a chaperone for this hydrophobic lipid that normally resides at the mid-plane of the membrane bilayer. The Coq3, Coq5, and Coq7 polypeptides are phosphorylated in a Coq8-dependent manner (shown by ‘???’). The function of Coq8 is still under investigation; although part of a family of atypical kinases, Coq8 has been shown to autophosphorylate, but not yet shown to phosphorylate any other proteins, in vitro or in vivo. It is speculated to have ATPase function and potentially has the ability to phosphorylate lipids or other small molecules. Hence the phosphorylation of Coq3, Coq5, and Coq7 may be from Coq8 or be produced via another kinase that is recruited to the CoQ synthome to act upon those particular polypeptides. In yeast, it has been shown that the phosphatase that dephosphorylates Coq7 is Ptc7s, the product of the spliced form of PTC7 (not shown).