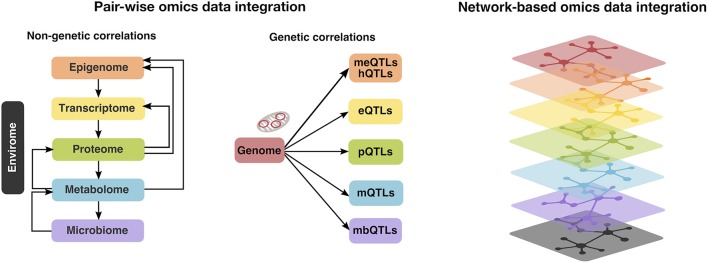
Figure 2.
Multi-omics (i.e., autosomal and mitochondrial genome, epigenome, transcriptome, proteome, metabolome, microbiome, and envirome) data integration can be conducted in two ways: pair-wise integrations, which can be further divided into non-genetic (left panel) and genetic correlations (middle panel). In the first case, one would examine the correlation patterns between the down-stream omics layers (e.g., metabolome and gut microbiome), whereas the second is achieved via the so called quantitative trait loci (QTL) mapping, linking genetic variation to methylation levels (meQTLs) or histone modifications (hQTLs), transcriptome (expression QTLs; eQTLs), protein (pQTLs), metabolite (mQTLs) or measures of microbiome (mbQTLs). Alternatively, multi-dimensional i.e., network-based integration approaches (right panel) exist, however their application in the context of CAD has so far been limited (22).