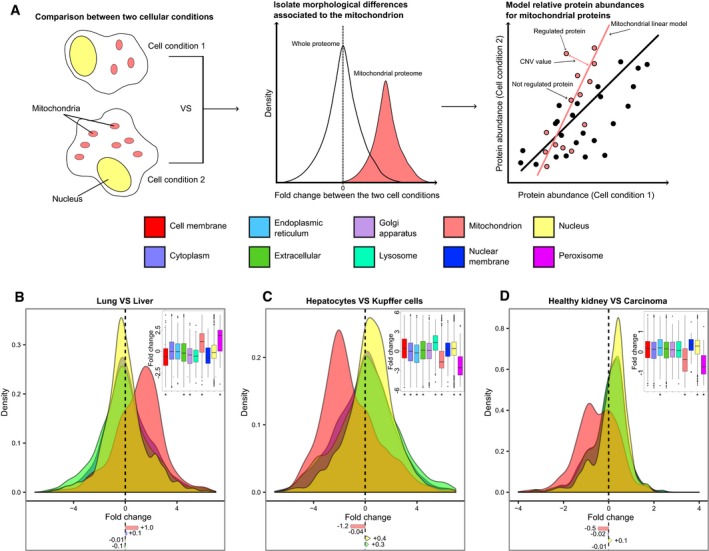
Analysis of protein abundance shift in cellular compartments in published datasets. Density plots for protein fold change distributions in different compartments for (B) lung vs. liver cells (Geiger
et al,
2013), (C) hepatocytes vs. Kupffer cells (Azimifar
et al,
2014), and (D) healthy human kidney vs. renal carcinoma cells (Guo
et al,
2015). Distribution of protein fold changes for 10 different compartments is shown as boxplot (inset); mean fold changes (log
2) are indicated below the density plots; below the boxplot, asterisks mark the cellular compartments that show significantly different distribution of fold changes compared to the whole proteome (Mann–Whitney test *
P < 0.01).