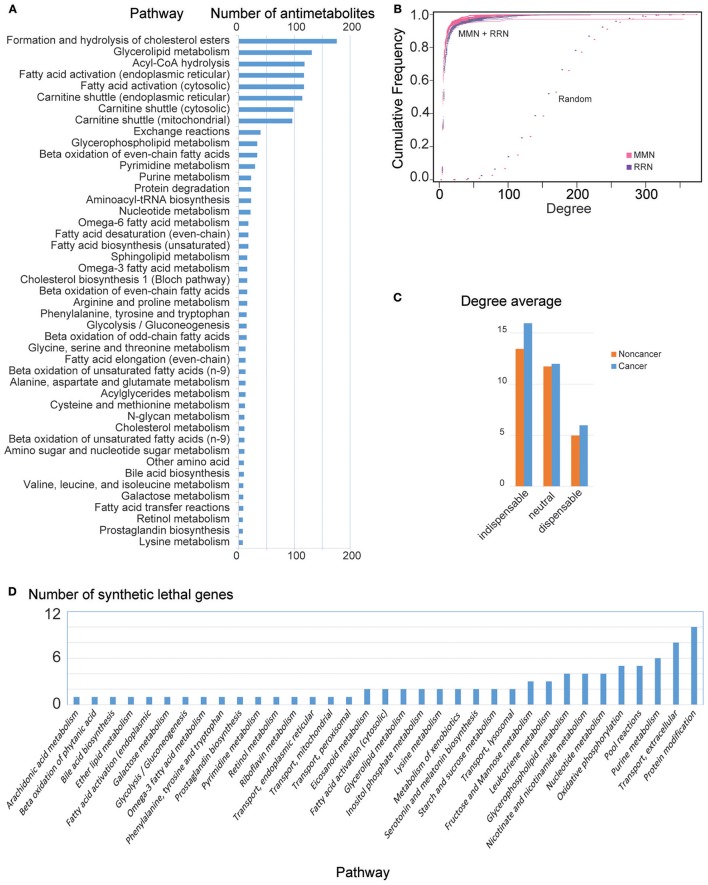
Figure 2.
General MMN and RRN node features in HCC. (A) Number of antimetabolites identified per pathway. Note that a predicted antimetabolite may be found in multiple pathways. (B) Cumulative degree distribution for MMN (purple) and RRN (magenta), compared to randomly generated models (Erdos–Rényi, dots). RRNs show qualitatively similar properties (results not shown). (C) Mean degree for indispensable, neutral, and dispensable metabolites in noncancer and HCC MMNs. RRNs show qualitatively similar properties (results not shown). (D) Number of genes leading to lethality identified per pathway. Note that a predicted synthetic lethal gene may be found in multiple pathways.