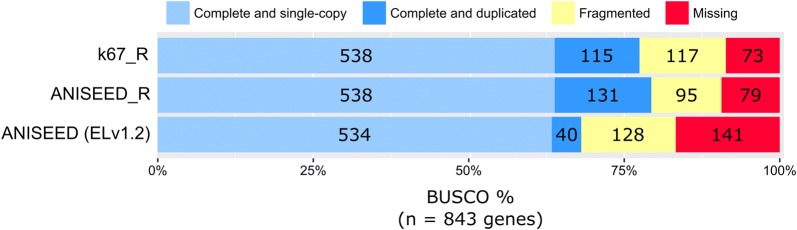
Fig. 1.
Benchmarking Universal Single-Copy Orthologs (BUSCO) in M. occidentalis. BUSCO was used to asses gene model predictions based on the previously published and released M. occidentalis genome assembly (ANISEED ELv1.2) and on the re-assemblies from this study (k67_R and ANISEED_R). BUSCO is used as a measure of completeness of gene model sets predicted from genome assemblies, searching for 843 highly conserved genes that should be present in single copies in > 90% of metazoan genomes. There are five categories of gene recovery: complete and single-copy, complete and duplicated, fragmented, and missing. Complete genes are those that preserve ~ 95% of the gene length. Single-copy genes are those with only one copy in the gene model set and duplicated genes are those with multiple copies identified either by gene duplication or assembly errors. Fragmented genes are those recovered with less than 95% of the gene length, and missing genes are those that are not found to be present at all