Fig. 5.
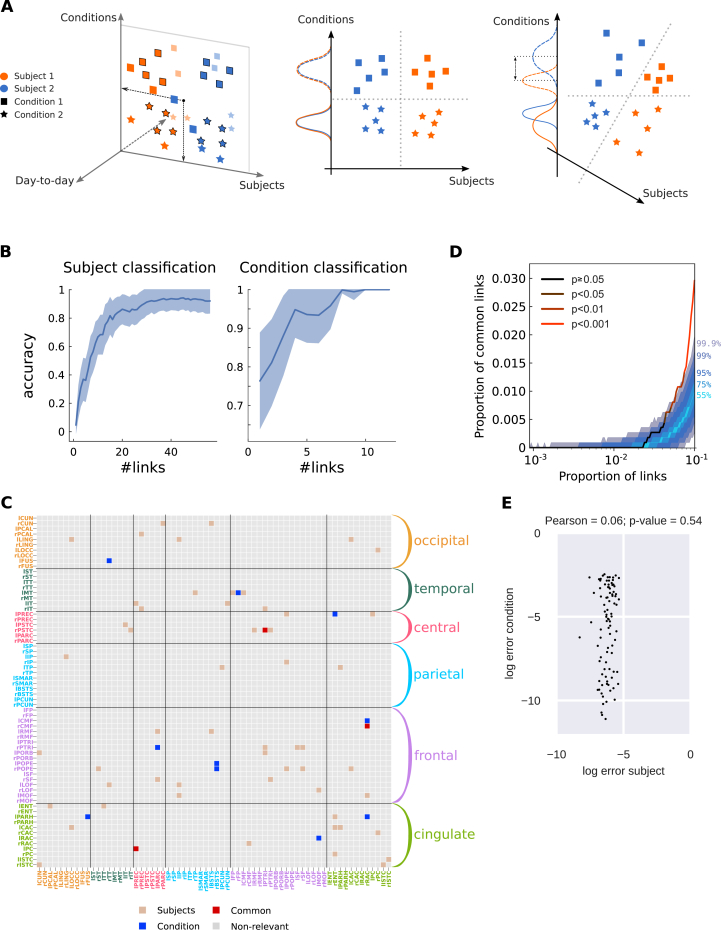
Twofold discrimination between subjects and conditions using EC. A) The left panel represents an idealized scheme of the twofold classification where the biomarkers for all sessions (colored symbols) are projected onto two subspaces, one for subjects (orange versus blue) and one for conditions (squares versus stars). The middle and right panels compare the situations where the subspaces (here axes) are orthogonal or not, respectively. In both cases, the dashed separatrices correspond to perfect classification. In the right panel, however, the condition “score” (projected value) mixes information about subjects, as indicated by the non-overlapping solid and dashed distributions. B) Performance of the classification for 19 subjects and 2 conditions using Dataset C as a function of number of links. Note the distinct scales for the y-axis, because the subject identification is a harder problem. C) Signatures of the most discriminative EC links (estimated with RFE, see text for details) for the twofold classification in B: 54 links for subject classification in brown, 10 for condition classification in blue, 3 common links in red. The ROIs are grouped in anatomical pools, as detailed in Supplementary Table S2. D) Proportion of common links between the subject and condition signatures as a function of selected links (in the order of the RFE ranking). Color coding is the same as in Fig. 4B: the two signatures are significantly different, i.e., with a number of common links corresponding to the null hypothesis with p-value (cf. legend) with up to 4% of the total links. E) Error of the subject and condition classifiers using the links of the support networks in C. The plotted dots represent the 95 sessions. The classifier errors are not correlated, as indicated by the Pearson correlation and corresponding p-value.