Figure 1.
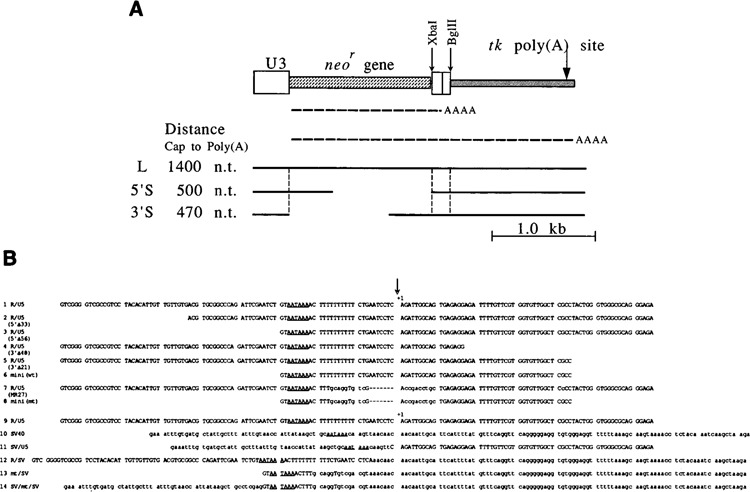
Structures of vectors and sequences used as the poly(A) signals. A. Vectors used. These vectors contained the U3 region of SNV for a promoter ( ), the neomycin resistance gene [neo
r] (
), the neomycin resistance gene [neo
r] ( ) to extend the distance from the cap site to the first poly(A) site, the first poly(A) signal to be tested (
) to extend the distance from the cap site to the first poly(A) site, the first poly(A) signal to be tested ( ), and the herpes simplex virus thymidine kinase [tk] gene to provide the second poly(A) site (
), and the herpes simplex virus thymidine kinase [tk] gene to provide the second poly(A) site ( ). The RNAs expressed from these vectors are shown by (----AA); one RNA species is polyadenylated at the first poly(A) site, and the other RNA species is polyadenylated at the tk poly(A) site. Tests for RNA 3′ end formation were performed using three types of vectors: L, 5′S, and 3′S. Gaps in the 5′S and 3′S constructs indicate deleted regions in these vectors. The distances from the cap site to the first poly(A) site in the L, 5′S, and 3′S constructs were approximately 1400, 500, and 470 nucleotides, respectively. By measuring levels of the two RNA species, the efficiency of RNA 3′ end formation was evaluated for the first poly(A) signals. A probe to detect RNA species transcribed from L and 5′S was the 5′ neor gene fragment. A probe to detect RNA species transcribed from L and 3′S was the 3′ neor gene fragment. (See Materials and Methods.) Other symbols: Xba I and Bgl II, sites of restriction enzymes used; vertical broken lines, boundaries of regions; thick lines, structures of vectors. B. Mutations of the SNV R/U5 region used to identify additional sequences necessary for RNA 3′ end formation. The R region is upstream from the poly(A) site, and the U5 region is downstream from the poly(A) site. The AATAAA poly (A) signal sequences are underlined. The poly(A) sites are marked by vertical arrows and +1. All the sequences were aligned at the poly(A) sites. The lowercase letters in the sequences indicate the nucleotides different from the parental sequence (R/U5). The broken lines in the sequences indicate the deleted nucleotides compared to the parental sequence. This R/U5 sequence contained two extra nucleotides at the 3′ end, GA, to create a Bgl II site, compared with the original R/U5 sequence. The addition of these nucleotides did not influence RNA 3′ end formation at the SNV poly(A) site (data not shown). The remainder of the vector sequences was identical for all constructs, except for pR/U5(3′Δ48). pR/U5(3′Δ48) contained 6 extra bases, CAGGTA BspM I site, at the junction between the U5 and tk sequences. These extra nucleotides did not influence RNA 3′ end formation (data not shown).
). The RNAs expressed from these vectors are shown by (----AA); one RNA species is polyadenylated at the first poly(A) site, and the other RNA species is polyadenylated at the tk poly(A) site. Tests for RNA 3′ end formation were performed using three types of vectors: L, 5′S, and 3′S. Gaps in the 5′S and 3′S constructs indicate deleted regions in these vectors. The distances from the cap site to the first poly(A) site in the L, 5′S, and 3′S constructs were approximately 1400, 500, and 470 nucleotides, respectively. By measuring levels of the two RNA species, the efficiency of RNA 3′ end formation was evaluated for the first poly(A) signals. A probe to detect RNA species transcribed from L and 5′S was the 5′ neor gene fragment. A probe to detect RNA species transcribed from L and 3′S was the 3′ neor gene fragment. (See Materials and Methods.) Other symbols: Xba I and Bgl II, sites of restriction enzymes used; vertical broken lines, boundaries of regions; thick lines, structures of vectors. B. Mutations of the SNV R/U5 region used to identify additional sequences necessary for RNA 3′ end formation. The R region is upstream from the poly(A) site, and the U5 region is downstream from the poly(A) site. The AATAAA poly (A) signal sequences are underlined. The poly(A) sites are marked by vertical arrows and +1. All the sequences were aligned at the poly(A) sites. The lowercase letters in the sequences indicate the nucleotides different from the parental sequence (R/U5). The broken lines in the sequences indicate the deleted nucleotides compared to the parental sequence. This R/U5 sequence contained two extra nucleotides at the 3′ end, GA, to create a Bgl II site, compared with the original R/U5 sequence. The addition of these nucleotides did not influence RNA 3′ end formation at the SNV poly(A) site (data not shown). The remainder of the vector sequences was identical for all constructs, except for pR/U5(3′Δ48). pR/U5(3′Δ48) contained 6 extra bases, CAGGTA BspM I site, at the junction between the U5 and tk sequences. These extra nucleotides did not influence RNA 3′ end formation (data not shown).
