Figure 4.
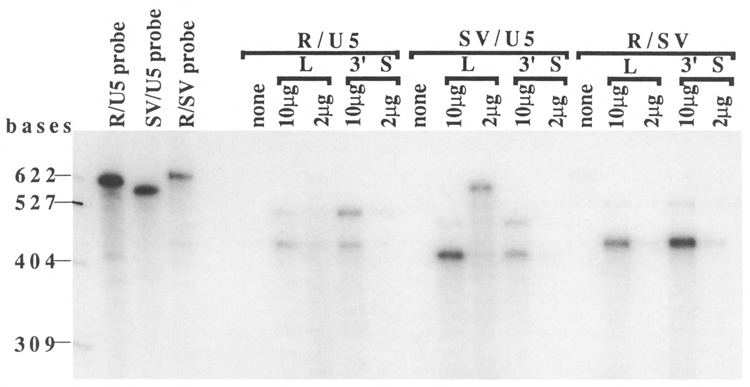
RNA protection assay. The sizes of marker RNAs are shown at the left. The input probes are shown in the left three lanes in this gel: R/U5, SV/U5, and R/SV probes. The names of probes and the names and amounts of the RNA samples are indicated at the top. The names of probes used were the same as the RNA samples (e.g., the R/U5 probe was used for RNA samples with R/U5 in the L and 3′S constructs). The sizes of the input RNA probes were 588 bases for the R/U5 probe, 560 bases for the SV/U5 probe, and 608 bases for the R/SV probe. Protected fragments were 433 bases (polyadenylated RNA) and 497 bases (pass-through RNA) at the R/U5 poly(A) site, 405 bases (polyadenylated RNA) and 469 bases (pass-through RNA) at the SV/U5 poly(A) site, and 440 bases (polyadenylated RNA) and 517 bases (pass-through RNA) at the R/SV poly(A) site. The RNA protection assay was performed as described by Farnham et al. (1985). Total RNA in reactions was adjusted to 10 μg with tRNA. “None” indicates reactions with only tRNA. RNA probes were synthesized in vitro using T7 RNA polymerase.
