Figure 3.
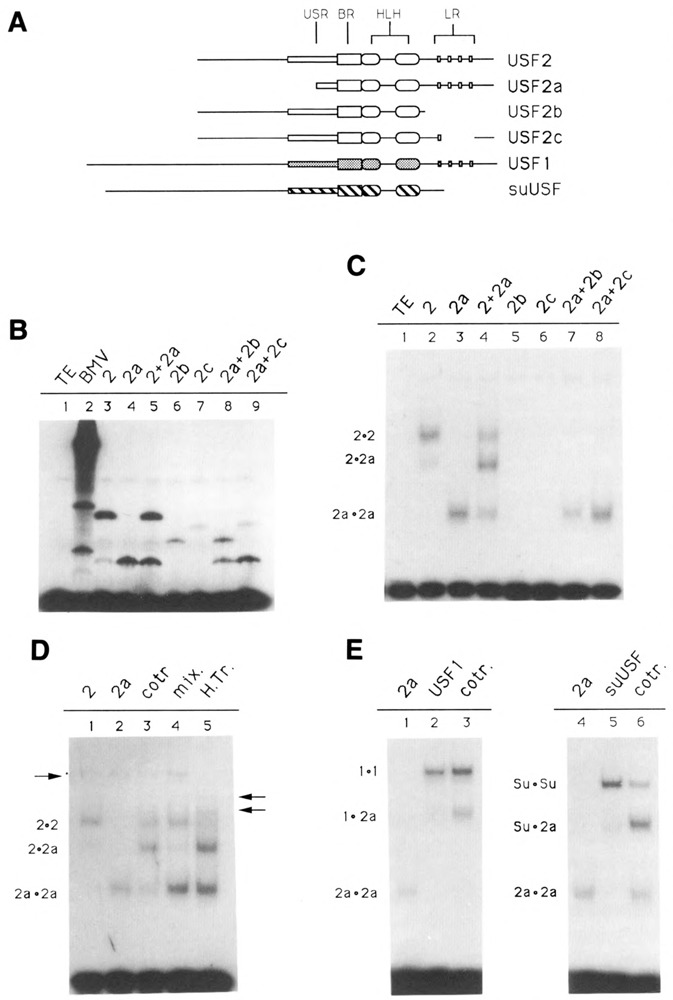
Domains of USF2 required for DNA-binding and dimer formation. A. Schematic of the various USF proteins. Shown are the USF proteins encoded by the pBS/ET-USF2, pBS/ET-USF2a, pBS/ET-USF2b, and PBS/ET-USF2c constructs (see Materials and Methods), which contain respectively amino acids 15–234, 103–234, 15–184, and 15–195/221–234 of hUSF2-C. The human USF1 (amino acids 1–310) and sea urchin USF (amino acids 1–265) were encoded by clones dI2 (Gregor et al., 1990) and 436–1 (Kozlowski et al., 1991), respectively. B. SDS gel analysis of in vitro translation products. Aliquots of reticulocyte lysates programmed with RNA derived from the various USF2 subclones, as indicated above each lane, were analyzed for the presence of translation products by SDS-PAGE. In lanes 5, 8, and 9, two different proteins were cotranslated. C. Localization of the minimum region of USF2 required for DNA-binding. The in vitro translated proteins shown in B were analyzed for DNA-binding activity by EMSA, using as a probe an oligonucleotide containing a consensus USF binding site (see Materials and Methods). The migrations of the different USF2 dimers are indicated at left. Longer exposure of the gel failed to detect any binding activity in the case of the USF2 LR mutants b and c (lanes 5 and 6). D. Formation of USF2 homodimers. USF2, or the N-terminal truncated USF2a mutant, were either translated separately (lanes 1, 2, 4, and 5) or cotranslated (lane 3), and the resulting DNA-binding species analyzed by EMSA. In lane 4, the individual proteins were simply mixed during the binding reaction. In lane 5, the two proteins were mixed, then heated for 7 minutes at 65°C prior to incubation with the oligonucleotide probe. The arrow at left indicates the migration of endogenous rabbit reticulocyte USF. The two arrows at right indicate the migration of heterodimers between rabbit USF and the human USF2 and USF2a proteins. E. Formation of USF heterodimers. The ability of USF2 to heterodimerize with human USF1 (lane 3) as well as with the sea urchin USF (lane 6) was investigated by cotranslating these proteins with the N-terminally truncated USF2a and analyzing the resulting DNA binding species by EMSA. Control reactions (lanes 1, 2, 3, and 4) were used to determine the migration of the individual homodimers, as indicated.
