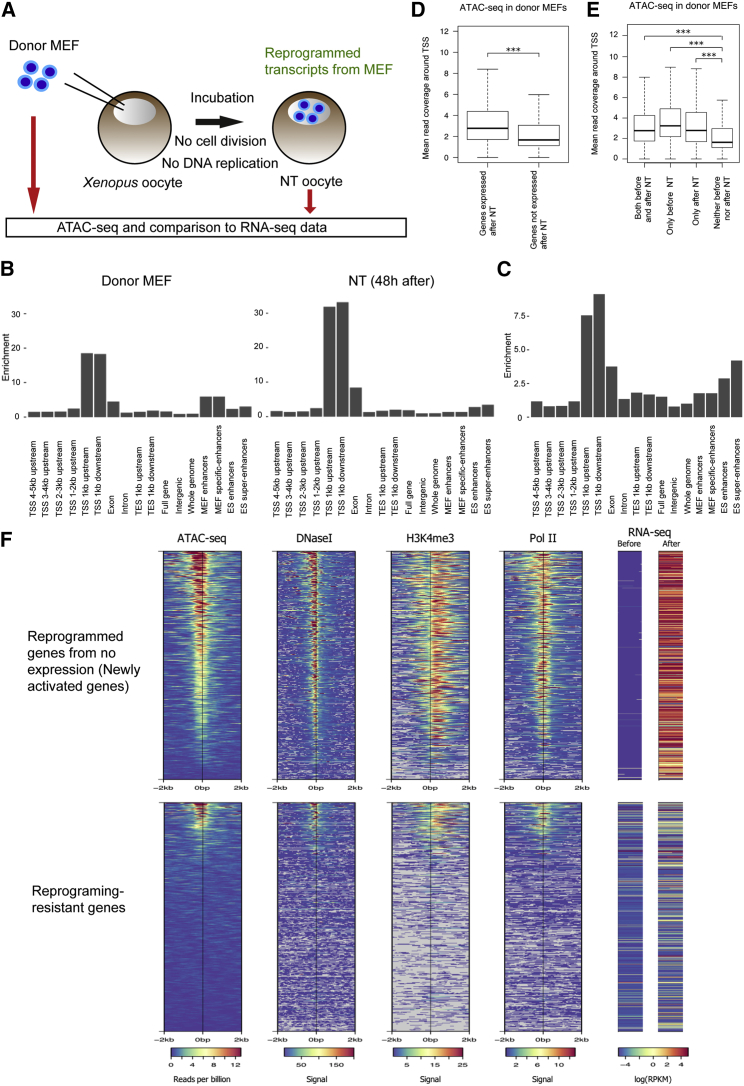
Figure 2.
Genes with Open TSSs Are Preferentially Reprogrammed upon NT to Xenopus laevis Oocytes
(A) MEFs are transplanted into the nuclei of Xenopus oocytes, reprogramming their transcription. MEFs before NT and reprogrammed MEFs were used for ATAC-seq. Two biologically independent NT experiments were performed for the subsequent analyses (10 NT oocytes, equivalent to 3,000 cells, were pooled in each experiment).
(B) The genomic distribution of ATAC-seq peaks representing open chromatin before and after NT. The y axis represents the enrichment of peaks in each type of genomic region relative to the whole genome.
(C) The genomic distribution of newly appeared ATAC-seq peaks after NT.
(D) ATAC-seq reads in donor MEFs were compared around TSSs. Genes were divided into two categories: expressed in NT oocytes and not expressed in NT oocytes. The y axis represents the mean read coverage in a 1-kb window centered on the TSS.
(E) ATAC-seq reads around TSSs in donor MEFs were compared among different gene categories: genes expressed before and after NT, those expressed only before NT, those expressed only after NT, and those expressed at neither time point.
(F) Representation of signal associated with open chromatin (ATAC-seq, DNase-seq, H3K4me3 ChIP-seq, and Pol II ChIP-seq) at TSSs of MEF genes reprogrammed in Xenopus oocytes. RNA-seq results (Jullien et al., 2014, Jullien et al., 2017) are shown at the right panel. Accession numbers for the DNase-seq, H3K4me3, and Pol II data are GSM1014172, GSM769029, and GSM918761, respectively.
∗∗∗p < 1E−6 by the Mann-Whitney U test.