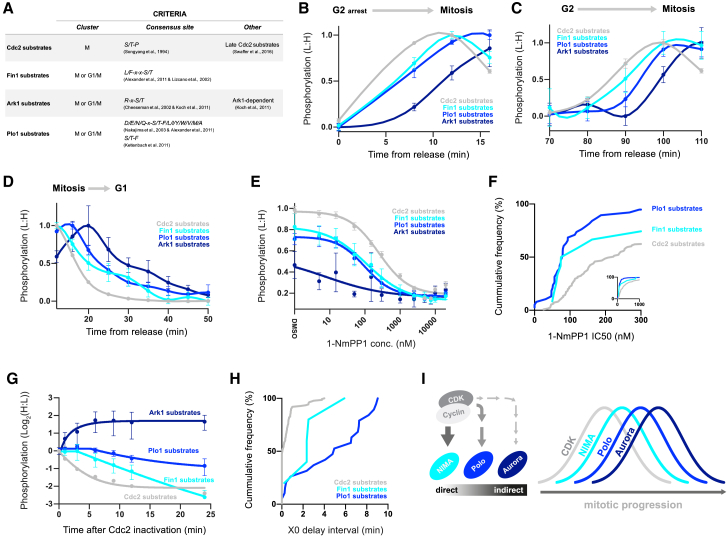
Figure 3.
The Dynamics of Kinase-Specific Phosphorylation during Mitosis
(A) The criteria used to define Cdc2 (CDK) and putative Fin1 (NIMA-related kinase), Plo1 (Polo-like kinase), and Ark1 (Aurora kinase) substrate sites during mitosis (see Experimental Procedures for details). Cdc2 substrates presented in this figure are only for those phosphorylated in mitosis defined as late substrates by Swaffer et al. (2016).
(B–D) The normalized mean (± SEM) phosphorylation (L:H) of Cdc2, Fin1, Plo1, and Ark1 substrate sites during mitotic entry (B and C) and mitotic exit (D). Mean (± SEM) values between 0 and 60 min (B and D) or 50 and 130 min (C) were normalized to the smallest (set to 0.0) and the largest (set to 1.0) values. Spline connects points. See Figures 1C and S1A for experimental design and details.
(E) The normalized mean (± SEM) phosphorylation (L:H) of Cdc2, Fin1, Plo1, and Ark1 substrate sites at different 1-NmPP1 concentrations. For Cdc2, Fin1, and Plo1, means (± SEMs) were calculated from phosphosites that could be fitted to a four-parameter logistic function (see Experimental Procedures for details). No Ark1 substrate sites could be fitted to the model. Means (± SEMs) were normalized to the largest mean during the cell cycle (set to 1.0). Curves are a four-parameter logistic function fit to the means. See Figure S1D for experimental design.
(F) The cumulative frequency of 1-NmPP1 IC50 values for individual Cdc2, Fin1, and Plo1 substrate sites, calculated from a four-parameter logistic function fit to the data. 1-NmPP1 IC50 values are plotted only for phosphosites that could be fitted to the function (see Experimental Procedures for details). No Ark1 substrate sites could be fitted to the function. Median IC50 values are 222, 97, and 79 nM for Cdc2, Fin1, and Plo1 substrate sites, respectively.
(G) The normalized mean (± SEM) phosphorylation (H:L) of Cdc2, Fin1, Plo1, and Ark1 substrate sites after Cdc2 inactivation in mitosis. For Cdc2, Fin1, and Plo1 substrates, means (± SEMs) were calculated from phosphosites that could be fitted to a plateau followed by one-phase decay function (see Experimental Procedures for details). No Ark1 substrate sites could be fitted to the function. Means (± SEMs) were normalized so that H:L [0 min] = 1.0. A plateau followed by one-phase decay fit to the data is shown. See Figure S1E for experimental design.
(H) The cumulative frequency of X0 values for individual Cdc2, Fin1, and Plo1 phosphosites. X0 is the delay interval between Cdc2 inactivation and the start of phosphosite dephosphorylation, calculated by fitting a plateau followed by one-phase decay function to the data. X0 values are plotted only for phosphosites fitted to the model (see Experimental Procedures for details). No Ark1 substrate sites could be fitted to the function. Median X0 values are 0.1, 2.4, and 5.0 min for Cdc2, Fin1, and Plo1 substrate sites, respectively.
(I) Schematic of the proposed model for the ordering of mitotic phosphorylation. Different mitotic kinases phosphorylate their substrates at different times during mitosis. The activation and inactivation timing is orchestrated, at least in part, by the differential dependence of mitotic kinases on upstream CDK activity.