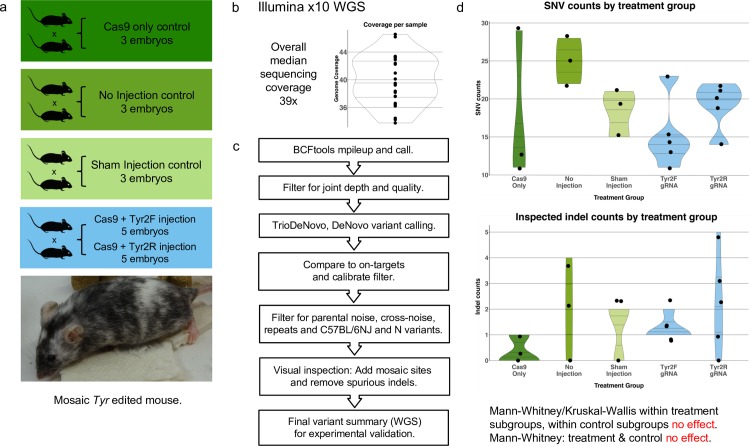
Fig 1. Analysis of CRISPR off-targets by whole genome sequencing.
(a) Experimental design: Four sets of C57BL/6N parents gave rise to 9 control embryos (3 “no injection”, 3 “sham injection” with water only and 3 “Cas9 only”), and 10 treated embryos (5 were injected with Cas9 and Tyr2F gRNA and 5 were injected with Cas9 and Tyr2R gRNA). (b) Whole genome sequencing: All 27 mice / embryos were subjected to whole genome sequencing with median depth 39.5x and an average of 3.4% of bases with read depth less than 11x. (c) Variant calling and filtering: starting from the joint variant call (bcftools mpileup + bcftools call), a sequence of filter steps were performed to detect de novo mutations and remove likely false positives arising from low-level parental mosaicism and alignment errors at repeat regions. Parental-noise: alternate-allele reads present in either parent. Cross-noise: alternate reads from all other (non-parental) samples. (d) Filtered SNV and Indel counts are not significantly different within control groups, within treatment groups, or between control and treatment groups.