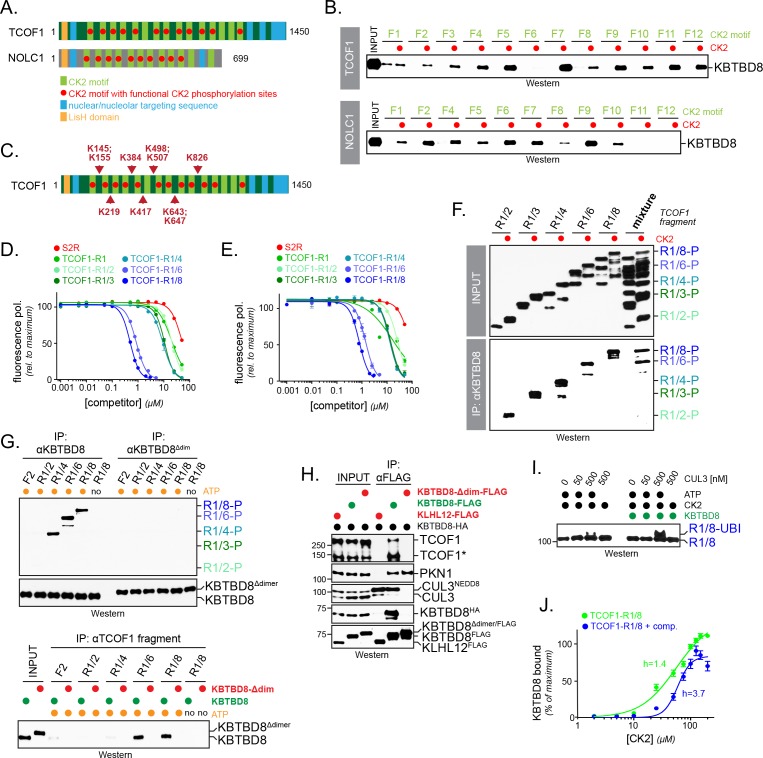
Figure 5. Multisite dependency of substrate recognition by CUL3KBTBD8 in vitro.
(A) TCOF1 and NOLC1 contain multiple CK2 motifs (light green). Functional motifs are marked with a red dot (non-functional motifs typically contain Arg substitutions that are incompatible with recognition by CK2). The LisH domain is a protein interaction module found in many proteins, but often with unknown function. (B) Multiple CK2 motifs are recognized by KBTBD8. TCOF1 or NOLC1 fragments containing single CK2 motifs (number according to position in sequence) were incubated with buffer or with CK2/ATP, immobilized on beads, and incubated with recombinant KBTBD8. Bound KBTBD8 was visualized by western blotting using a specific antibody. (C) TCOF1 is ubiquitylated in vivo on multiple Lys residues that are all in close proximity to CK2 motifs. Ubiquitylated TCOF1 was purified under denaturing conditions from 293 T cells and ubiquitylated Lys residues were determined by mass spectrometry. The figure depicts the location of ubiquitylated lysine residues identified in individual peptides. (D) The more CK2 motifs are in a substrate, the better it binds to KBTBD8. A single phosphorylated and fluorescently labeled CK2 motif of TCOF1 was bound to KBTBD8. Unlabeled TCOF1 competitor fragments containing either two (R1/2), three (R1/3), four (R1/4), six (R1/6) or eight (R1/8) CK2 motifs were phosphorylated and titrated into the binding reaction, and dissociation of the reporter peptide was monitored by loss of fluorescence polarization. (E) The increase in affinity afforded by multiple CK2 motifs also applies to monomeric KBTBD8. A monomeric variant of KBTBD8 was produced by mutation of conserved residues at the BTB dimer interface, as described for KLHL12 (McGourty et al., 2016). Binding of a single phosphorylated CK2 motif of TCOF1 to monomeric KBTBD8 was then monitored in the presence of competitor peptides by fluorescence polarization, as described above. (F) Multiple CK2 motifs increase substrate affinity for KBTBD8. TCOF1 fragments containing either two (R1/2), three (R1/3), four (R1/4), six (R1/6) or eight (R1/8) CK2 motifs were incubated with immobilized KBTBD8 either in the presence or absence of CK2. In the last two lanes, all TCOF1 fragments were mixed and incubated at the same time with KBTBD8. Binding was monitored by western botting (top lanes: input; bottom lanes: KBTBD8-immunoprecipitation). (G) Multiple CK2 motifs increase substrate affinity towards KBTBD8 at low substrate concentrations. Upper panel: 450 nM of TCOF1 fragments with a single (F2) or multiple (R1/2, R1/4, R1/8) CK2 motifs were incubated with either wild-type KBTBD8 or monomeric KBTBD8Δdim, and KBTBD8 was then affinity-purified using a specific antibody. Lower panel: Same binding reaction as above, but TCOF1 was immobilized on beads. Complex formation was analyzed by western blotting. (H) Dimerization of KBTBD8 is required for TCOF1, but not PKN1, binding in cells. Cells expressing indicated KBTBD8 variants were subjected to KBTBD8FLAG affinity-purification and analyzed for dimerization (KBTBD8HA), CUL3-binding, or substrate recognition (TCOF1, PKN1) using Western blotting with specific antibodies. (I) CUL3KBTBD8-substrates with multiple CK2 motifs are monoubiquitylated. Ubiquitylation of TCOF1-R1/8, a fragment containing eight CK2 motifs, was analyzed in vitro using purified neddylated CUL3-RBX1, KBTBD8, E1, UBE2D3, ubiquitin and ATP, as described above. (J) Ultrasensitivity of the KBTBD8-TCOF1 interaction. Binding of TCOF1-R1/8 to KBTBD8 was analyzed in the presence of increasing CK2 concentrations, as described in the Figure 5F. Western blots of three independent experiments were quantified by Image J, and error bars denote standard deviation. Green curve: binding reaction in the absence of competitor; blue curve: binding of TCOF1-R1/8 in the presence of a ten-fold excess of TCOF1-R1/2, which mimics endogenous CK2 substrates that typically have one or two CK2 motifs.
Figure 5—figure supplement 1. N- and C-terminal TCOF1 fragments both require multiple CK2 motifs for recognition.