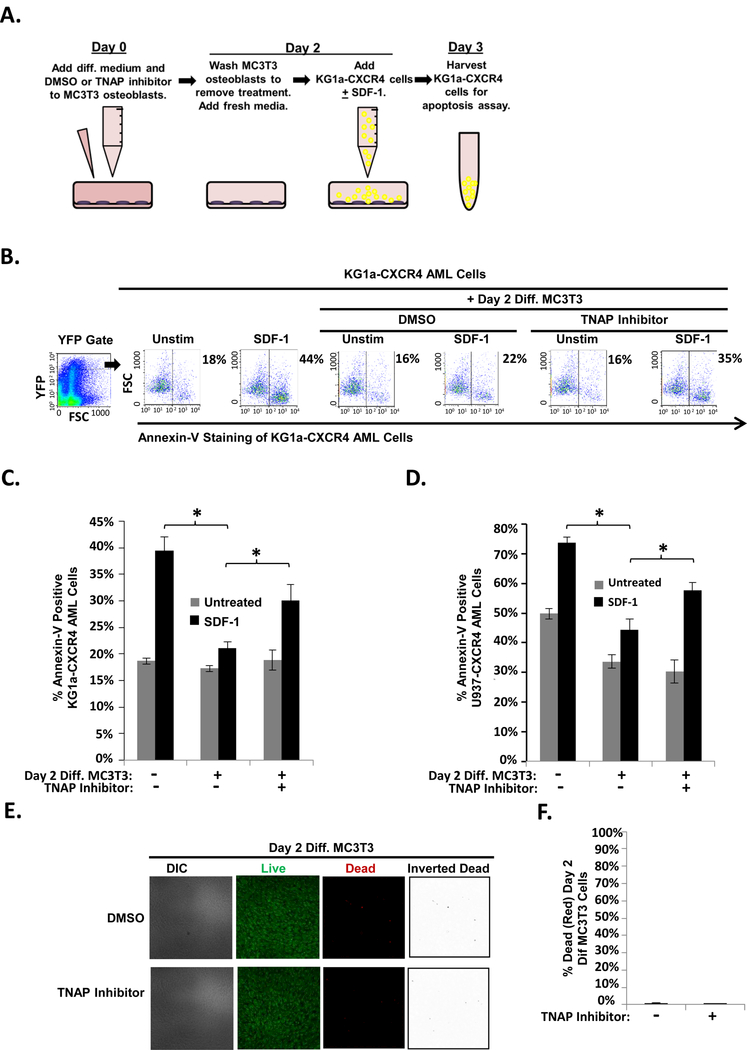
FIGURE 5. TNAP inhibition reduces MC3T3 osteoblast-mediated protection of KG1a-CXCR4 and U937-CXCR4 AML cells from SDF-1-induced apoptosis.
(A) Depiction of MC3T3 osteoblast and KG1a-CXCR4 AML cell line co-culture model. On Day 0, MC3T3 osteoblasts were treated with osteogenic differentiation media and either 0.1% DMSO or 10μM TNAP inhibitor MLS-0038949. On Day 2, the osteoblasts were washed to remove the DMSO or TNAP inhibitor pretreatment and were then co-cultured with KG1a-CXCR4 AML cells. The indicated wells were challenged with 1.3 × 10−8 M SDF-1. On Day 3, the KG1a-CXCR4 AML cells were harvested and assayed for apoptosis utilizing annexin-V staining and flow cytometry. (B) A representative experiment as performed in (A) shows the percentage of annexin-V positive KG1a-CXCR4 cells from each culture within the YFP gate indicated (gating on cells with high levels of CXCR4 expression as the CXCR4 is YFP tagged). (C) Summary of multiple experiments performed as in (A). Bars depict mean results + S.E.M., n=3; *, indicates p<0.05. (D) Summary of multiple experiments performed as in (A) except with U937-CXCR4 AML cells instead of KG1a-CXCR4 AML cells. Bars depict mean results + S.E.M., n=3; *, indicates p<0.05. (E) MC3T3 cells were treated as in Fig. 4a except live/dead staining and imaging occurred on Day 2 (without the addition of KG1a-CXCR4 AML cells, AMD3100, or SDF-1). Live/dead staining and confocal imaging of live (green) and dead (red) cells were used to ensure MC3T3 cell viability in the presence of TNAP inhibitor. Images were acquired on three separate days for a total of 15 images analyzed for each condition. (F) Statistical summary of (F). Bars depict mean results + S.E.M., n=3.