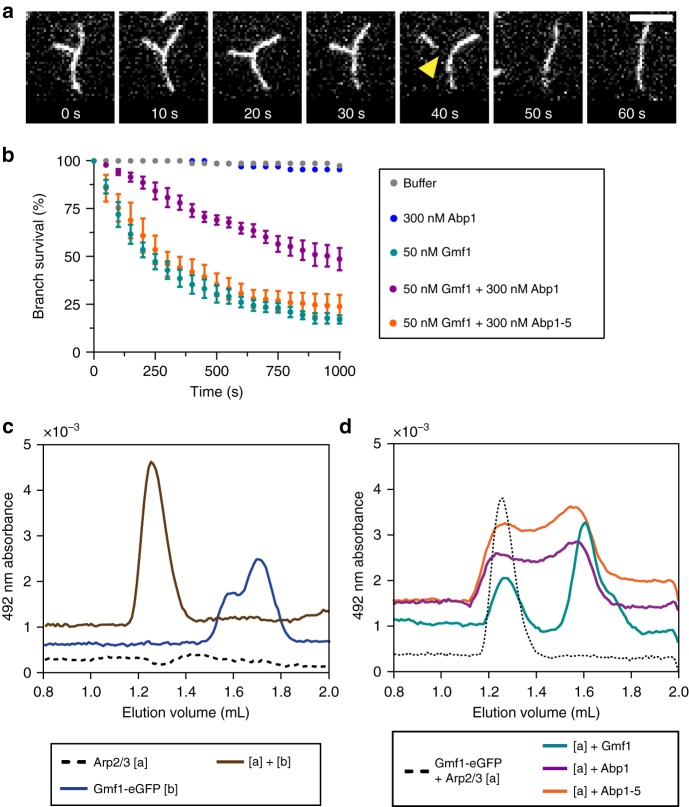
Fig. 5.
Abp1 protects actin filament branches from GMF-induced debranching. a Time points from a TIRF microscopy debranching assay, with yellow arrow marking debranching event. Branched filaments were first polymerized and biotin-anchored in the TIRF chamber with actin (10% OG-labeled, 0.5% biotin-labeled), Arp2/3 complex, and GST-VCA. Then, proteins of interest were flowed in, and effects on branch stability were monitored over time. Scale bar, 5 µm. b Cumulative survival curves of branches in the presence of different proteins, quantified from n = 50 branches in each of four independent trials per condition. For the two negative controls (buffer alone, and 300 nM Abp1), data are from a single trial. Error bars, s.e.m. c Size exclusion chromatography profiles for the migration of 0.8 µM Gmf1-eGFP, monitored by absorbance at 492 nm, in the presence (brown line) and absence (blue line) of 1 µM Arp2/3 complex. The dotted line shows that Arp2/3 complex alone (1 µM) has no appreciable signal at 492 nm. d Size exclusion chromatography competition assay for Arp2/3 complex binding, following the absorbance of the Gmf1-eGFP absorbance at 492 nm. The dotted line shows the migration of Gmf1-eGFP (0.8 µM) in the presence of Arp2/3 complex (1 µM), and the colored lines show how the migration of Gmf1-eGFP with Arp2/3 complex is affected by the further addition of binding competitors: 5 µM unlabeled Gmf1 (cyan line), 1 µM Abp1 (magenta line), and 1 µM Abp1-5 (orange line). Chromatograms are vertically offset for clarity