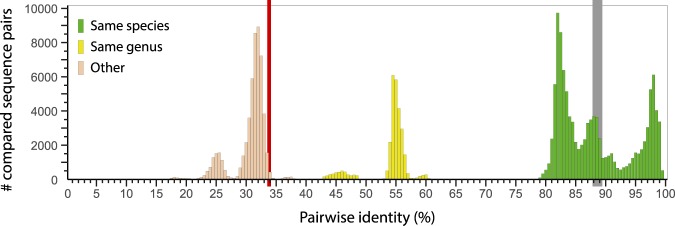
Figure 3.
Pairwise sequence comparison analysis of arterivirus genomes, including the newly discovered three complete genome sequences of Olivier’s shrew virus 1 (OSV-1) variants A–C, using the NCBI PASC tool (https://www.ncbi.nlm.nih.gov/sutils/pasc)18. Pairwise similarity between the three sequences ranges from 87.89–89.39% (marked in grey), indicating all three sequences belong to viruses that ought to be classified in the same species. Pairwise similarity with the closest related virus, porcine reproductive and respiratory syndrome virus 1 (PRRSV-1), ranges from 33.49–34.01% (marked in red), indicating the need for the establishment of a new arterivirus genus and species. Color coding was modified to accurately represent the current taxonomic organization of the family Arteriviridae, using 39–41% and 71–77% as genus and species cut-offs, respectively.