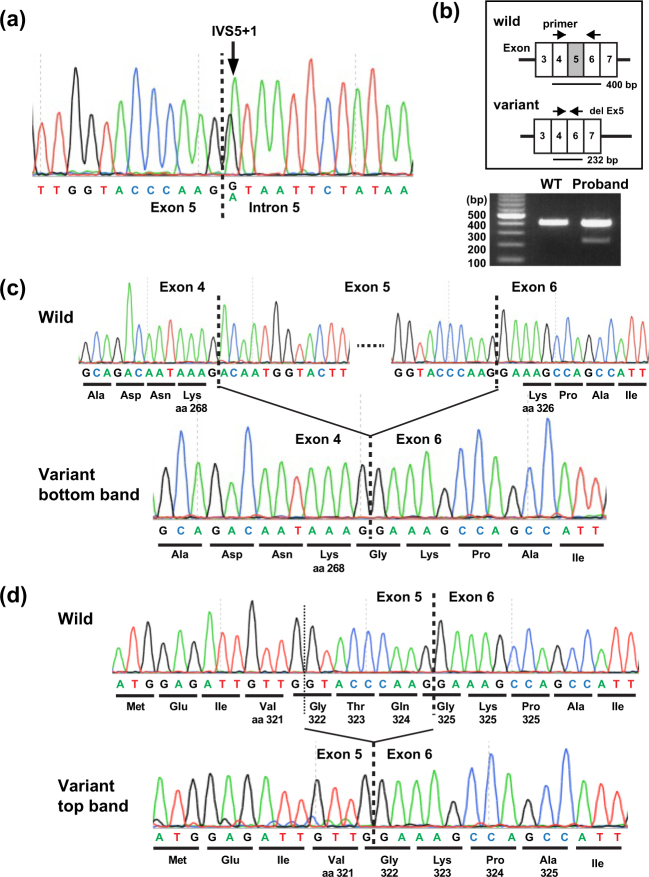
Fig. 2.
TGFBR1 variant analysis. (a) Genomic DNA sequencing revealed a heterozygous single-base substitution (c.973 + 1 G > A) in the proband. (b–d) Splice analysis of exon 5. (b) Arrows indicate the position of PCR (top panel). Wild-type and variant cDNA were predicted to produce 400 bp and 232 bp fragments. The actual PCR products on 1.2% agarose gel (bottom panel) in a healthy unaffected volunteer (left, WT) and the proband (right). The doublet bands in the gel indicate the splicing abnormality. (c, d) Direct sequencing of cDNA from wild-type (top) and variant (bottom) alleles in the proband revealed a 168 bp deletion corresponding to entire exon 5 skipping (c) and a last 9 bp deletion of exon 5 (d). aa amino acid position of TGFBR1