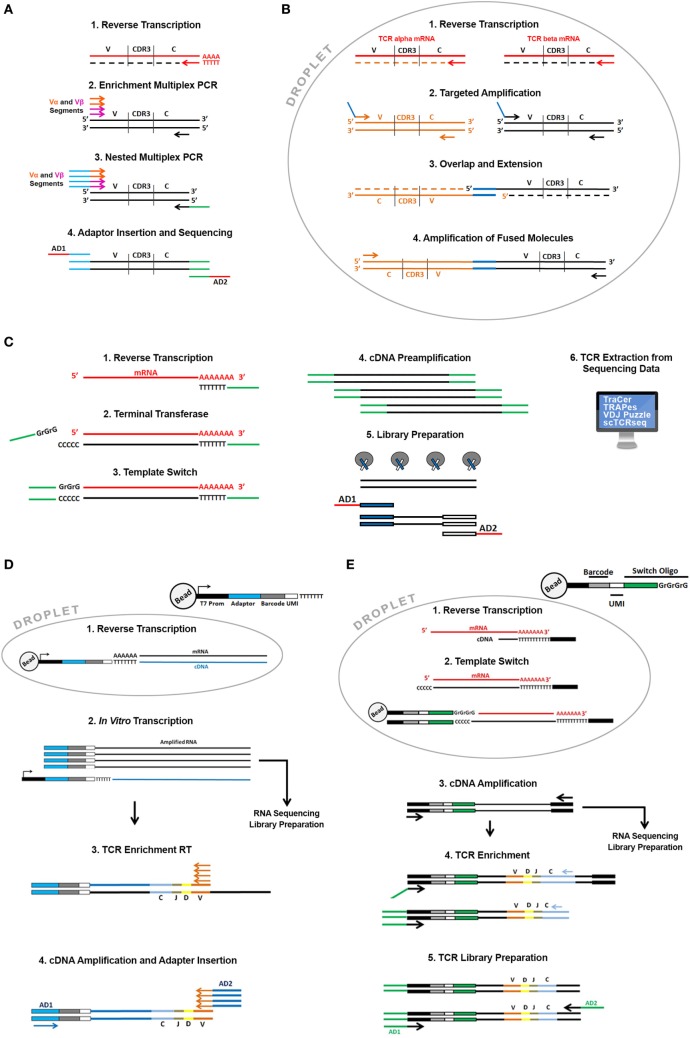
Figure 2.
Overview of the available single cell T cell receptor (TCR) sequencing approaches. Direct TCR enrichment and sequencing. In (A) single cell TCR transcripts are enriched by a multiplex PCR performed after RT reaction using a pool of forward primers spanning all the annotated productive V alpha and V beta fragments and reverse primers designed on the constant region of alpha and beta chains. Barcoded adaptors then are added by PCR enabling pooling and sequencing by Next-Generation Sequencing. In (B) cells are captured in water-in-oil droplets using microfluidic emulsion-based devices along with specific RT and PCR reagents. In each droplet, TCR alpha and beta transcripts of a single cell are specifically reverse transcribed with RT primers designed on the constant region of alpha and beta chain. cDNA is successively amplified using a pool of forward primers designed on all the alpha and beta segments and reverse primers designed on the constant region. Alpha and beta primers contain overlapping sequences at their 5′ends that enable the synthesis of TCR alpha and beta fusion sequences through an overlap-extension mechanism. Fused molecules are pooled breaking the emulsion and further enriched by a nested amplification and sequenced. TCR reconstruction from single cell “full length” RNA sequencing (RNA-seq) data. (C) Single cells sorted in plates or captured using microfluidic devices are lysed, and total mRNA is reverse transcribed with an oligo dT primed reaction. Through a template-switch mechanism a universal sequence is added at the 5′ end of the transcript. This sequence, shared with the dT primer used in the RT reaction is then used to amplify cDNA before library preparation. In the library preparation step, full-length cDNA is “tagmented” using Transposase and Tag sequences inserted by transposase are then used to amplify cDNA and to insert barcoded sequencing adaptors (AD1 and AD2). Libraries are then sequenced and TCR sequences can be extracted from all transcriptome using dedicated bioinformatics algorithms (TraCer, TraPes, VDJ Puzzle). Pairing Single cell TCR sequencing and RNA-seq using emulsion-based protocols. (D) Thousands of cells in parallel are partitioned into oil-in-water droplets. After a lysis step their mRNA is reverse transcribed using a pool of specific RT primers containing the same “cell barcode” used to tag the cell transcriptome, a Unique Molecular Identifier (UMI). UMI is different for each primer enabling the digital counting of mRNA transcripts and sequencing of the T7 promoter. cDNA is then amplified by in vitro transcription. After amplification barcoded RNAs are pooled and processed together. Amplified RNA is then used as template to enrich for TCR sequences and to generate RNA-seq libraries according to the InDrop protocol. During RNA-seq library preparation RNA is fragmented and only the 3′ end of transcripts is sequenced. For TCR enrichment amplified RNA is reverse transcribed using a pool of RT primers spanning V alpha and V beta segments and then amplified using “internal” V alpha and beta primers and primers designed on the constant regions. During this PCR reaction sequencing adaptors are also added (AD1 and AD2). (E) Thousands of cells in parallel are partitioned into oil-in-water droplets. After a lysis step their mRNA is reverse transcribed using an oligo dT primer. Through a template-switch mechanism, a primer containing a cell barcode and a UMI is added at the 5′ end of the transcript. After RT reaction droplets are broken and cDNAs pooled and amplified using external primer designed on dT and switch oligonucleotides, respectively. Amplified full-length cDNA is then used as template to enrich TCR sequencing and and/or fragmented and processed to generate RNA-seq libraries. TCR enrichment is performed using nested PCR with a forward primer spanning the switch oligo and reverse primers designed on the constant region of alpha and beta chains. PCR products are then partially fragmented and sequencing adaptors are added (AD1 and AD2).