Fig. 2.
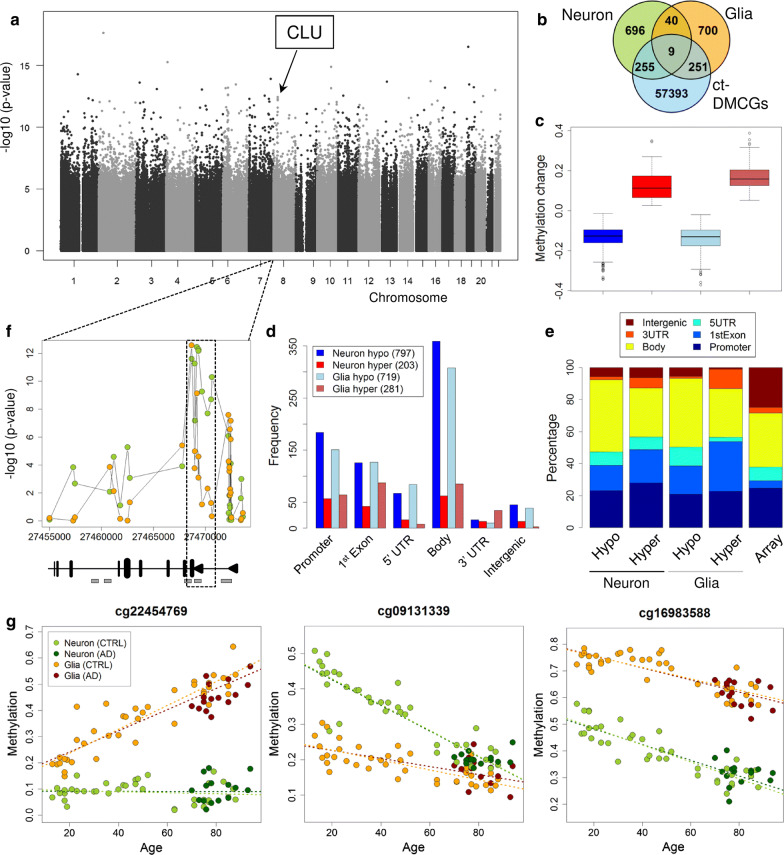
Aging analysis in neurons and glia. a Genomic p value distribution for the aging analysis in neurons. The arrow marks a region on chromosome 8 with a concentrated set of top-ranking CpGs (see f). b Overlap of top 1000 aging-DMCGs between neuron and glia, but approx. 25% of each set are ct-DMCGs. c Methylation change from 8 youngest to 8 oldest samples for top 1000 aging CpGs in neurons and glia, respectively. Boxes colored as in d. d Genomic region classification of top 1000 aging-DMCGs from neuron and glia, respectively. CpGs are classified in respect of hypo- or hypermethylation in 8 oldest versus 8 youngest CTRL samples. e Relative distribution of top 1000 aging-DMCGs in relation to array design. f p-value distribution at the clusterin gene locus (CLU) in the age analysis in neuron (green) and glia (red), respectively. A set of 10 adjacent CpGs (dashed square) shows strong age-association in healthy neurons (but only partially in glia). Numbers on x-axis give coordinates on chromosome 8 (hg19). Cartoon below illustrates the relative location of exons (black boxes), promoters (black triangles) and annotated sites of high DNaseI hypersensitivity (gray boxes) according to UCSC genome browser. g Age-dependent methylation changes that occur exclusively in one cell-type can lead to either emergence (left panel) or disappearance (mid) of significant differences between cell-types. Aging effects that are synchronized between cell-types are stable ct-DMCGs over lifetime (right). Adding AD samples leads to similar results (bright dashed lines: regression lines based on CTRLs, dark dashed lines: CTRLs + AD cases) (color figure online)