FIGURE 2.
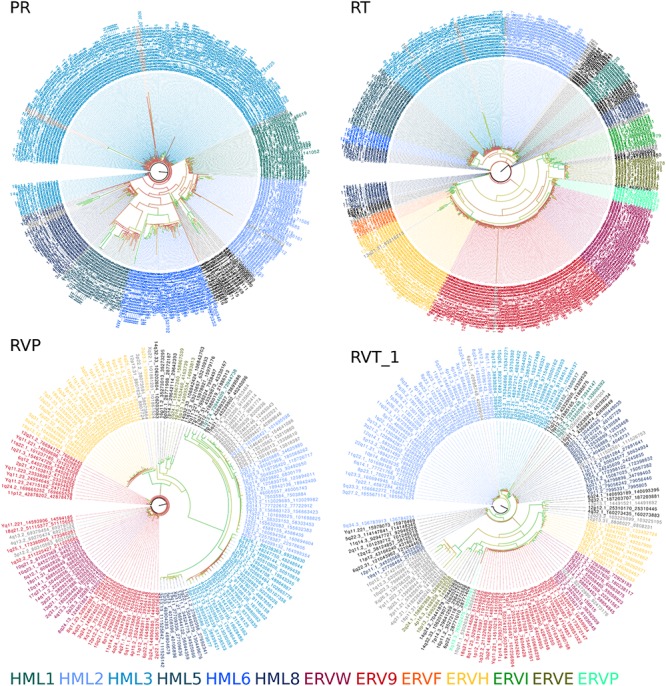
Amino acid sequence alignment derived phylogenies of human genomic endogenous retroviral sequences. A tree is shown for each search method (PR, protease BLAST; RT, reverse transcriptase BLAST; RVP, HMMER for RVP; RVT_1, HMMER for RVT_1). Leaves are colored by their classification through the evolutionary placement algorithm implemented by RAxML (see color legend at bottom of figure). Branch coloring represents bootstrap support, where red indicates poor support (closer to zero) and green indicates good support (closer to 100).
