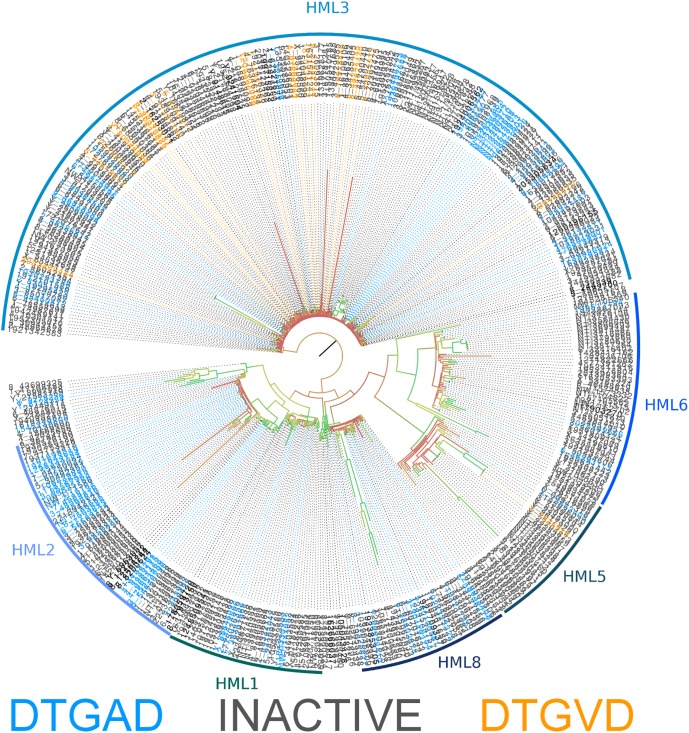
FIGURE 4.
Phylogeny of human genomic nucleotide sequences derived using BLAST and the ERVK HK2 PR. Potentially active sequences with a DTGAD motif in their active site are colored blue, and those with DTGVD are colored orange. Sequences containing inactivating mutations are colored gray. The classification of each area of the tree is indicated by the name associated with the arc around the perimeter. Branch coloring represents bootstrap support, where red indicates poor support (closer to zero) and green indicates good support (closer to 100). Placement of the root at the branch connecting HK3 is only a matter of visual convenience – the real root branch is more likely that leading to HK8. This alignment is available in Additional File 41.