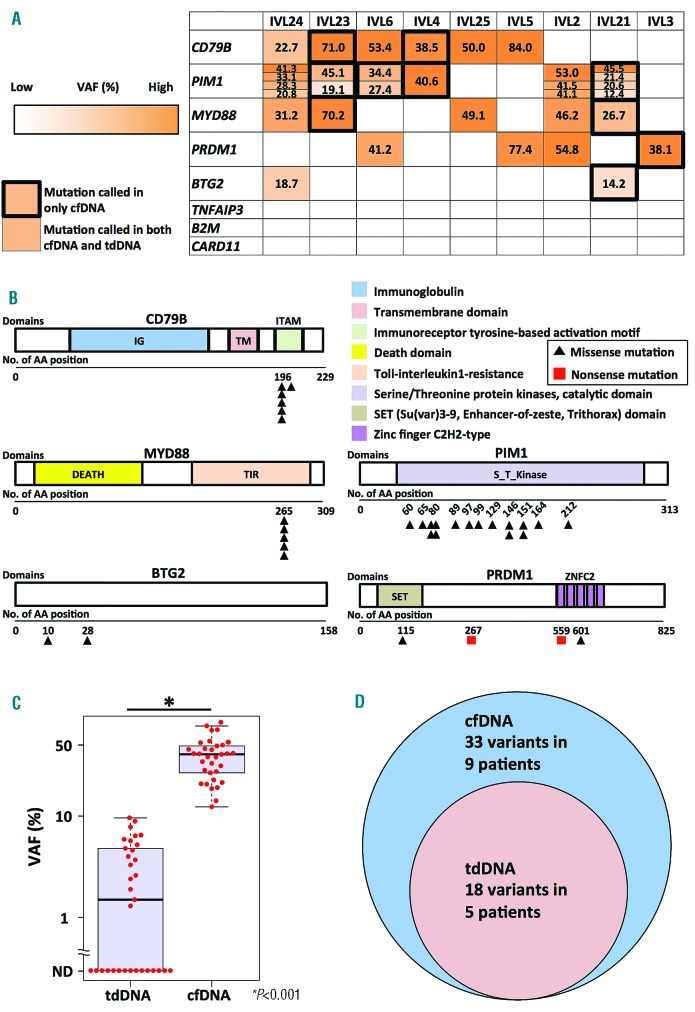
Figure 1.
Results of targeted sequencing in 9 patients using cell-free DNAs, tissue-derived DNAs, and reference genomic DNAs.A. Summary of gene alterations in 9 IVLBCL patients. Numbers in boxes represent variant allele frequencies (%) in cell-free DNAs (cfDNAs). Mutations found in only cfDNAs are surrounded by black thick outline. Variants are color-coded according to their variant allele frequencies of cfDNAs. VAF: Variant allele frequencies; cfDNA: cell-free DNA; tdDNA: tissue-derived DNA. B. The positions of mutations in each gene at protein levels. A black triangle indicates missense mutation. A red box indicates nonsense mutation. IG, Immunoglobulin; TM, Transmembrane domain; ITAM, Immunoreceptor tyrosine-based activation motif; DEATH, Death domain; TIR, Toll-interleukin1-resistance; S_T_Kinase, Serine/Threonine protein kinases, catalytic domain; SET, Su(var)3–9, Enhancer-of-zeste, Trithorax domain; ZNFC2, Zinc-finger C2H2-type; AA, Amino acid. C. Comparison of variant allele frequencies in tissue-derived DNAs with those in cell-free DNAs. VAF, Variant allele frequencies; tdDNA, tissue derived DNA; cfDNA, cell-free DNA. D. Venn diagram of variants called in tissue-derived DNAs with those in cell-free DNAs; tdDNA: tissue-derived DNA; cfDNA: cell-free DNA.