Figure 2.
Platelet Transcriptome Profiling in HIV
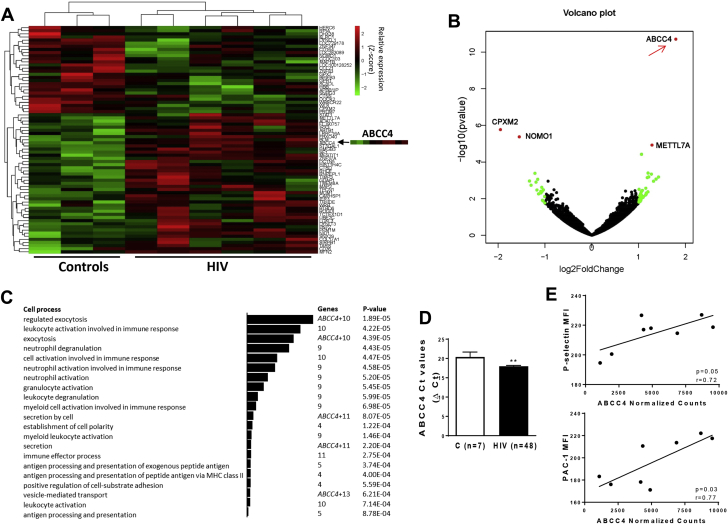
(A) Heat map from hierarchical clustering of differentially expressed genes between controls and HIV platelets. Genes with p < 0.01 were considered for the analysis. The ABCC4 heat map is highlighted. (B) The data for all genes are plotted as log2 fold change versus the log10 of the p value. The top genes that are significantly different (sorted by p value) are indicated. (C) GO enrichment analysis (2 unranked list) of differentially expressed genes (p < 0.01). Bars indicate significantly enriched GO terms associated with cellular process and cell function. (D) ABCC4 mRNA expression in platelets from healthy volunteers (n = 7) and HIV (n = 48) subjects. qPCR analysis was reported expressing the ABCC4 Ct values after normalization with 18S5 RNA. **p < 0.01. (E) ABCC4 expression (RNA-Seq normalized counts) correlates with surface P-selectin and PAC-1 (activated Integrin αIIbβ3) staining in persons with HIV and controls, measured by flow cytometry (r = 0.72; p = 0.05 and r = 0.77; p = 0.03, respectively). Ct = concentration-time product; MFI = median fluorescence intensity; MHC = major histocompatibility complex; PAC-1 = activated glycoprotein IIb/IIIa receptor; other abbreviations as in Figure 1.