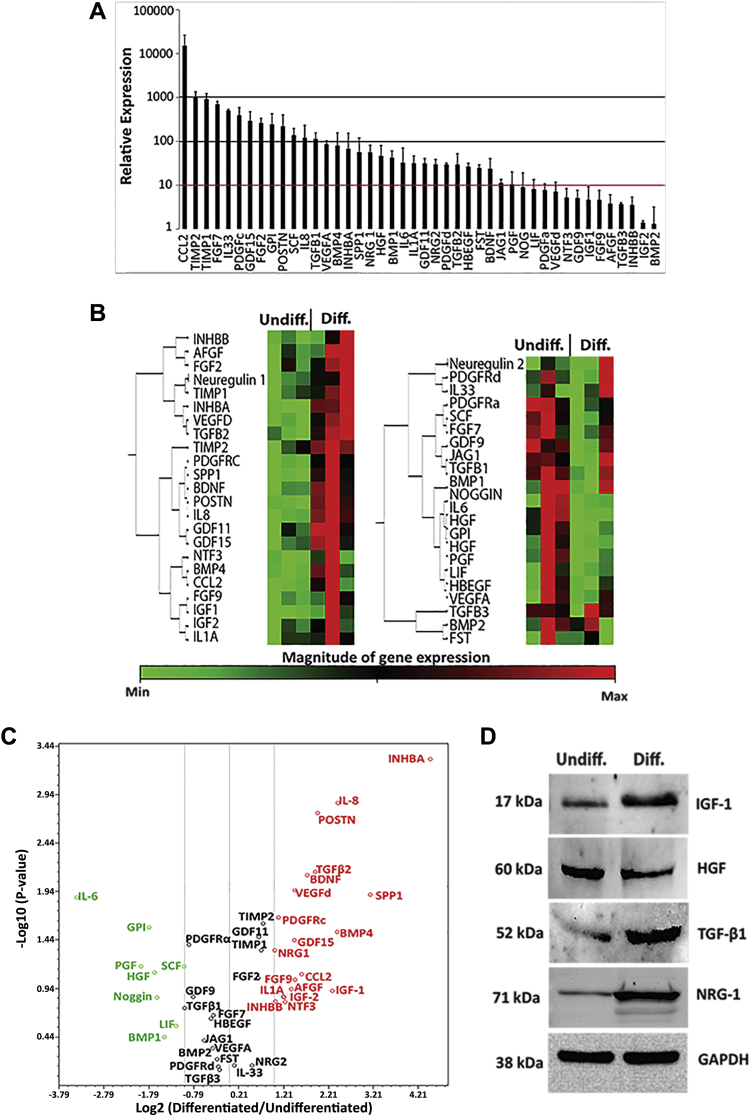
Figure 5.
Allogeneic pPICs Express an Array of Proregenerative Paracrine Factors
(A) Expression profile of pPICs arrayed by quantitative reverse transcription polymerase chain reaction, relative expression to average expression of 5 housekeeping genes ACTB, B2M, GAPDH, HPRT1, and RPL13A. Data represent mean ± SD, n = 3. (B) Hierarchical clustering of differentially regulated pPIC transcripts. The colors in ascending order from green to red represent the magnitude of gene expression for the measured average difference values. The tree on the left of the clustergram indicates the pairwise similarity relationships between the clustered expression patterns, n = 3 for both undifferentiated and differentiated pPICs (24 h in differentiation medium). (C) Genes that were differentially regulated by at least 2-fold were selected and are illustrated in a volcano plot. Red indicates those genes that were up-regulated, black represents genes whose expression levels did not change, whereas green indicates genes that were down-regulated in differentiated pPICs compared with undifferentiated pPICs, n = 3. (D) Western blots confirmed that pPIC exposure to a myogenic permissive environment led to an increase in IGF-1, TGF-β1, and NRG-1, and a decrease in HGF protein expression, n = 3. Diff = differentiated; pPIC = porcine PW1pos/Pax7neg interstitial cell; Undiff = undifferentiated.