FIGURE 2.
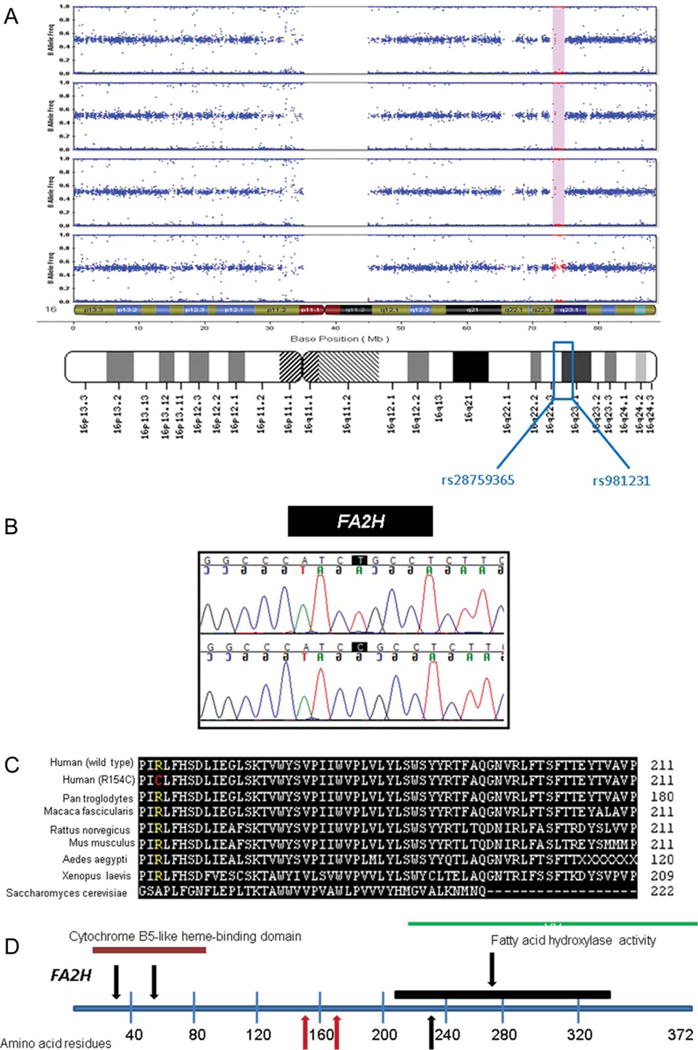
Identification and analysis of a novel FA2H mutation. (A) A shared block of homozygosity nearly 2 Mb in length, common to all three affected members of the index family but absent in the unaffected brother genotyped, was identified. (B) FA2H was selected as a prominent candidate gene given its critical role in lipid metabolism; sequencing identified a c.460C>T homozygous transition in all three affected family members. (C) By aligning homologs across species using ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html), amino acid sequence conservation across species was compared; the mutation identified (R154C) was predicted to alter a highly conserved arginine residue. (D) Comparison of this novel mutation with other identified mutations19,20 (red arrows = Family 1 and 2; black arrows = previously reported mutations; black line indicates extent of splice site mutation predicted to result in skipping of exons 5 and 6).
