Abstract
Cyclin F is a part of the Skp, Cullin, F-box containing ligase complex. The activity of cyclin F includes cell cycle control, centrosome duplication and response to DNA damage. The cyclin F expression pattern is very similar to cyclin A, but cyclin F is an orphan cyclin without its cyclin-dependent kinase partner. There is little evidence concerning the role of cyclin F in cancer. In the present study, for the first time, we present analysis from The Cancer Genome Atlas (TCGA) data in the context of expression of cyclin F mRNA in melanoma patients. Our original in silico analysis, not published elsewhere before, revealed that high expression of cyclin F in melanoma patients is associated with worse overall survival. Cyclin F and ribonucleotide reductase family member 2 (RRM2) compose a functional axis responsible for nucleotide metabolism. Impairment in this pathway may contribute to increased DNA damage repair and drug resistance. Additionally, we analyzed the expression of RRM2 mRNA and discovered that high expression of RRM2 is associated with worse overall survival. To shed more light on cyclin F overexpression in melanoma, we analyzed all protein data available in the TCGA melanoma dataset. It was found that in patients with upregulated cyclin F mRNA, we noted increased activity of pathways related to cell cycle and DNA damage repair. These data will support further in vitro and in vivo studies on the involvement of cyclin F in skin cutaneous melanoma.
Keywords: cyclin F, CCNF, ribonucleotide reductase, melanoma, skin cancer
Introduction
Although melanoma comprises 5% of all skin-related tumors, it is responsible for 75% of the deaths caused by this type of cancer. Although significant progress has been made in the last decade and the number of cases has significantly decreased, the overall mortality rate has remained steady. New treatment strategies based on BRAF inhibitors or CTLA-4 blocking antibodies have provided only slight benefit to patients with stage IV melanoma and melanoma metastases. This moderate success provides the rationale to continue research on expanding therapies focusing on cancer biology and targeting molecular pathways crucial for proliferation, metastasis and respond to treatment (1–3).
DNA synthesis and repair require coordinated deoxyribonucleoside triphosphate (dNTP) supply as basic building blocks. Impaired balance of the dNTP pool affects S phase duration time, DNA synthesis fidelity, as well as the ability and effectiveness of DNA repair. Loss of control over these processes can also trigger genome instability and may initiate cancerogenesis. The increased demand for deoxyribonucleotides is serviced by upregulation of ribonucleotide reductase (RNR), which reduces the 2′ carbon of a ribonucleoside diphosphate and has been considered as the rate-limiting step in dNTP production. RNR as a heterodimeric protein consists of three subunits – one ribonucleotide reductase family member 1 (RRM1) and two molecules of RRM2. While RRM1 expression is constant throughout the cell cycle, the expression of RRM2 fluctuates and peaks at S phase, when the need for nucleotide synthesis is the highest. The degradation of RRM2 occurs in late G2 phase of the cell cycle in the nucleus and is controlled by Skp, Cullin, F-box containing (SCF)cyclin F ubiquitin ligase complex. The SCF complex is composed of three proteins: Skp1 and Cul1, which provide a scaffold, and F-box protein, which is responsible for target recognition (4).
Cyclin F, like other cyclins, has both cyclin and F-box domains, but it does not bind or activate any known cyclin-dependent kinase (CDK). The expression profile of cyclin F is similar to cyclin A and fluctuates throughout the cell cycle. At the protein level, cyclin F appears in the S phase, peaks before M phase, and then its expression decreases dramatically. It is clearly visible that changes in the expression of cyclin F negatively correlates with the RRM2 level, which may suggest their cooperation in the axis, important for genome stability and DNA repair (5). As it has been suggested, overexpression of RRM2 is associated with poorer patient prognosis in melanoma and many other cancers. Furthermore, cells with high content of RRM2 are characterized by much more effective DNA repair systems which impair the effectiveness of therapy (6–9).
The aim of our in silico analysis was to take the first step in the elucidation of the precise mechanism of the cyclin F (CCNF)-RRM2 axis in skin melanoma. The study aims to accelerate the development and to inspire other scientific teams to conduct similar research in the field.
In the present study, using the data available in the cBioPortal database, we showed for first time that high expression of cyclin F mRNA is associated with poorer prognosis in patients with skin cutaneous melanoma. Additionally, we present an overview of the molecular pathways involved in the cell cycle, cell death and DNA repair which are activated differentially in patients who exhibit high and low expression of cyclin F and RRM2.
Materials and methods
Analysis of publicly available data
To assess the expression profile of cyclin F and RMM2 mRNA, we obtained data from The Cancer Genome Atlas via www.cBioPortal.org (10). Patients were divided into groups: with CCNF or RRM2 mRNA upregulated expression (z-score >0) and with downregulated mRNA expression (z-score ≤0) and then, for each mRNA, we conducted overall survival and disease-free survival analysis. The same source was used for protein level comparison in patients with upregulated and downregulated cyclin F and RRM2 mRNA. In turn, we analyzed obtained information and used Reactome (http://reactome.org) and ToppGene Suite (http://toppgene.cchmc.org) to organize data into biological processes and functional molecular pathways.
Statistical analysis
In the life span study of the melanoma patients, the data were analyzed with Kaplan-Meier survival analysis with included log-rank test for trend tests. Comparisons between groups expressing different levels of mRNA or proteins were conducted using Mann-Whitney U-test. All statistical analyses were performed using GraphPad Prism 7.0 (GraphPad Software, Inc., La Jolla, CA, USA).
Results
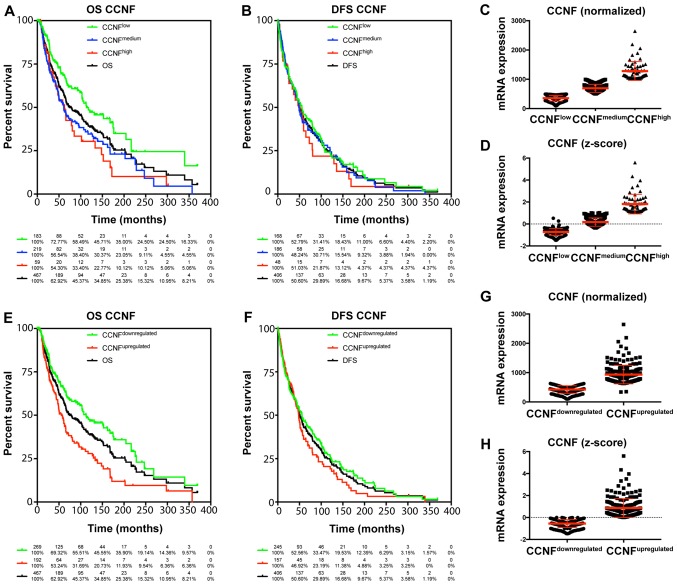
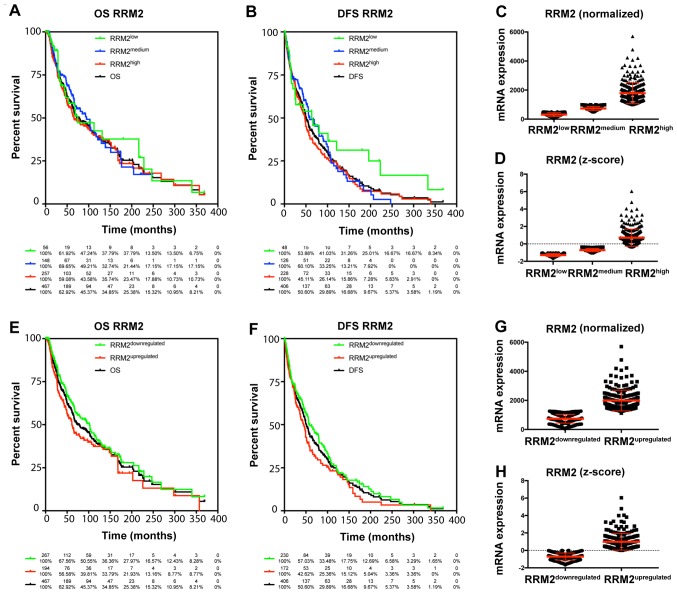
The TCGA data were used to characterize the prognostic value of cyclin F and RRM2 mRNA in melanoma. The results showed that increased expression of cyclin F mRNA is associated with worse outcome in melanoma patients (Fig. 1; Tables I and II). Median survival in patients with upregulated cyclin F was significantly lower (112.48 vs. 55.55 months; P<0.0001). No significance in disease-free survival (DFS) was found. Furthermore, expression of RRM2 mRNA had a significant influence on median survival (102.04 vs. 61.47; P=0.034), but no effect on DSF was noted (Fig. 2; Tables I and II). Cyclin F significantly altered the expression of different cellular proteins. The expression of proteins negatively and positively correlated with CCNF mRNA are listed in Tables III and V. Representative plots are shown in Figs. 3 and 4. Analogous data for RRM2 mRNA expression are shown in Tables VII and IX, and representative plots are presented in Figs. 5 and 6.
Figure 1.
(A, B, E and F) High expression of CCNF mRNA is associated with poorer prognosis in melanoma patients. Patients with melanoma were analyzed by Kaplan-Meier survival estimation (log-rank test). (C, D, G and H) Representative plots of patients with differential expression of CCNF mRNA: normalized (C and G) and z-score (D and H). OS, overall survival; DFS, disease-free survival; CCNF, cyclin F.
Table I.
Association of CCNF and RRM2 mRNA expression on the survival of melanoma patients.
| Overall survival (%) | Disease-free survival (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| Factor | Median survival (months) | Disease-free median survival (months) | 5 years | 10 years | 15 years | 5 years | 10 years | 15 years |
| Total | 74.67 | 51.08 | 58.79 | 39.20 | 25.38 | 42.85 | 24.93 | 12.09 |
| CCNF expression (normalized) | ||||||||
| CCNFlow | 113.44 | 55.49 | 68.40 | 48.55 | 35.00 | 46.05 | 24.17 | 15.40 |
| CCNFmedium | 61.10 | 48.59 | 52.24 | 34.64 | 23.05 | 41.18 | 26.87 | 10.88 |
| CCNFhigh | 62.75 | 51.08 | 51.44 | 30.36 | 10.12 | 36.45 | 21.87 | 4.37 |
| CCNF expression (z-score) | ||||||||
| CCNFdownregulated | 112.48 | 55.85 | 65.83 | 47.57 | 35.90 | 46.50 | 27.27 | 15.15 |
| CCNFupregulated | 55.55 | 48.00 | 48.06 | 27.93 | 11.93 | 36.14 | 20.46 | 6.50 |
| RRM2 expression (normalized) | ||||||||
| RRM2low | 74.67 | 63.40 | 58.48 | 42.51 | 37.79 | 53.88 | 36.47 | 31.26 |
| RRM2medium | 94.91 | 58.97 | 64.07 | 39.11 | 21.44 | 49.07 | 22.17 | 10.57 |
| RRM2high | 65.83 | 47.60 | 55.55 | 39.11 | 23.47 | 37.74 | 24.46 | 8.74 |
| RRM2 expression (z-score) | ||||||||
| RRM2downregulated | 102.04 | 58.97 | 63.15 | 41.44 | 27.97 | 49.34 | 26.30 | 15.51 |
| RRM2upregulated | 61.47 | 44.15 | 52.73 | 37.40 | 21.93 | 34.77 | 23.16 | 6.72 |
CCNF, cyclin F; RRM2, ribonucleotide reductase family member 2.
Table II.
Changes in overall survival and disease-free survival as associated with CCNF and RRM2 mRNA expression in melanoma patients.
| Overall survival | Disease-free survival | |||||||
|---|---|---|---|---|---|---|---|---|
| Factor | HR | 95% CI | P-value | Significance | HR | 95% CI | P-value | Significance |
| CCNF expression (normalized) | ||||||||
| CCNFlow vs. total | 0.73 | 0.57–0.93 | 0.0119 | * | 0.96 | 0.77–1.19 | 0.6915 | NS |
| CCNFmedium vs. total | 1.21 | 0.96–1.54 | 0.1070 | NS | 1.01 | 0.81–1.27 | 0.9072 | NS |
| CCNFhigh vs. total | 1.33 | 0.90–1.97 | 0.1576 | NS | 1.11 | 0.75–1.62 | 0.6087 | NS |
| CCNFlow vs. CCNFmedium | 0.60 | 0.45–0.80 | 0.0005 | *** | 0.95 | 0.73–1.27 | 0.6733 | NS |
| CCNFlow vs. CCNFhigh | 0.48 | 0.30–0.77 | 0.0022 | ** | 0.86 | 0.57–1.30 | 0.4748 | NS |
| CCNFmedium vs. CCNFhigh | 0.94 | 0.64–1.39 | 0.7717 | NS | 0.92 | 0.61–1.37 | 0.6784 | NS |
| CCNF expression (z-score) | ||||||||
| CCNFdownregulated vs. total | 0.79 | 0.63–0.98 | 0.0317 | * | 0.94 | 0.78–1.15 | 0.5671 | NS |
| CCNFupregulated vs. total | 1.42 | 1.11–1.82 | 0.0053 | ** | 1.11 | 0.87–1.40 | 0.3980 | NS |
| CCNFdownregulated vs. CCNFupregulated | 0.54 | 0.43–0.75 | <0.0001 | **** | 0.85 | 0.66–1.10 | 0.2211 | NS |
| RRM2 expression (normalized) | ||||||||
| RRM2low vs. total | 0.87 | 0.59–1.30 | 0.5052 | NS | 0.77 | 0.53–1.12 | 0.1693 | NS |
| RRM2medium vs. total | 0.93 | 0.71–1.22 | 0.5970 | NS | 0.96 | 0.75–1.24 | 0.7756 | NS |
| RRM2high vs. total | 0.95 | 0.76–1.17 | 0.6165 | NS | 1.08 | 0.88–1.32 | 0.4596 | NS |
| RRM2low vs. RRM2medium | 1.10 | 0.68–1.76 | 0.7059 | NS | 1.31 | 0.85–2.03 | 0.2260 | NS |
| RRM2low vs. RRM2high | 0.84 | 0.56–1.27 | 0.4156 | NS | 0.73 | 0.50–1.08 | 0.1134 | NS |
| RRM2medium vs. RRM2high | 0.88 | 0.66–1.18 | 0.3845 | NS | 0.90 | 0.69–1.17 | 0.4161 | NS |
| RRM2 expression (z-score) | ||||||||
| RRM2downregulated vs. total | 0.88 | 0.71–1.09 | 0.2507 | NS | 1.11 | 0.91–1.36 | 0.3133 | NS |
| RRM2upregulated vs. total | 1.17 | 0.92–1.50 | 0.1960 | NS | 1.15 | 0.92–1.44 | 0.2233 | NS |
| RRM2downregulated vs. RRM2upregulated | 0.75 | 0.57–0.98 | 0.0344 | * | 0.78 | 0.61–1.00 | 0.0529 | NS |
HR, hazard ratio; CI, confidence interval; CCNF, cyclin F; RRM2, ribonucleotide reductase family member 2. ****, extremely significant (P<0.0001); ***, extremely significant (P=0.0001 to 0.001); **, very significant (P=0.001 to 0.01); *, significant (P=0.01 to 0.05); NS, not significant (P≥0.05).
Figure 2.
(A, B, E and F) High expression of RRM2 is associated with less favorable outcome in melanoma patients. Patients with melanoma were analyzed by Kaplan-Meier survival estimation (log-rank test). (C, D, G and H) Representative plots of patients with differential expression of RRM2: normalized (C and G) and z-score (D and H). OS, overall survival; DFS, disease-free survival; RRM2, ribonucleotide reductase family member 2.
Table III.
Expression of proteins which are negatively correlated with CCNF.
| CCNFdownregulated | CCNFupregulated | ||||
|---|---|---|---|---|---|
| RPPA (z-score) | |||||
| Protein | Gene | upregulated | downregulated | P-value | Significance |
| A-Raf_pS299 | ARAF | 0.0567 | −0.0105 | 0.0279 | * |
| Annexin_VII | ANXA7 | 0.0085 | −0.0491 | 0.0055 | ** |
| Annexin-1 | ANXA1 | 0.2359 | −0.0402 | 0.0006 | *** |
| AR | AR | 0.0662 | −0.0072 | 0.0380 | * |
| Axl | AXL | 0.1741 | −0.0276 | 0.0283 | * |
| Bak | BAK1 | 0.0059 | −0.0199 | 0.5374 | NS |
| Bcl-2 | BCL2 | 0.0461 | −0.1069 | 0.0190 | * |
| Bcl-xL | BCL2L1 | 0.0578 | −0.0127 | 0.0609 | NS |
| Bim | BCL2L11 | 0.0081 | −0.1046 | 0.0200 | * |
| Caveolin-1 | CAV1 | 0.2809 | −0.0344 | 0.0013 | ** |
| CD31 | PECAM1 | 0.0548 | −0.0108 | 0.0260 | * |
| CD49b | ITGA2 | 0.1129 | −0.0100 | <0.0001 | **** |
| Chk1_pS345 | CHEK1 | 0.0011 | −0.0009 | 0.6241 | NS |
| DJ-1 | PARK7 | 0.0503 | −0.0112 | 0.0743 | NS |
| EGFR_pY1068 | EGFR | 0.0817 | −0.0107 | 0.0015 | ** |
| ER-α | ESR1 | 0.0900 | −0.0314 | 0.0002 | *** |
| FOXO3a | FOXO3 | 0.0724 | −0.0102 | <0.0001 | **** |
| GATA3 | GATA3 | 0.0186 | −0.0356 | 0.0287 | * |
| GATA6 | GATA6 | 0.0949 | −0.0295 | 0.0132 | * |
| HER2 | ERBB2 | 0.0678 | −0.0827 | 0.0036 | ** |
| HER3 | ERBB3 | 0.0023 | −0.0620 | 0.2038 | NS |
| HER3_pY1289 | ERBB3 | 0.0086 | −0.0137 | 0.1953 | NS |
| INPP4B | INPP4B | 0.0761 | −0.0258 | 0.0008 | *** |
| JAB1 | COPS5 | 0.0558 | −0.1180 | <0.0001 | **** |
| JNK2 | MAPK9 | 0.0404 | −0.0589 | 0.0083 | ** |
| Myosin-IIa | MYH9 | 0.0003 | −0.0030 | 0.9509 | NS |
| p27 | CDKN1B | 0.0582 | −0.1027 | <0.0001 | **** |
| p38_pT180_Y182 | MAPK14 | 0.0115 | −0.0346 | 0.3252 | NS |
| p53 | TP53 | 0.0557 | −0.0223 | 0.0021 | ** |
| PARP_cleaved | PARP1 | 0.0227 | −0.0241 | 0.0773 | NS |
| PDCD4 | PDCD4 | 0.0854 | −0.1255 | 0.0025 | ** |
| PEA15 | PEA15 | 0.0238 | −0.0052 | 0.4346 | NS |
| PI3K-p110-α | PIK3CA | 0.0097 | −0.0625 | 0.0315 | * |
| PKC-α | PRKCA | 0.1358 | −0.2574 | <0.0001 | **** |
| PKC-α_pS657 | PRKCA | 0.1951 | −0.1638 | <0.0001 | **** |
| PKC-δ_pS664 | PRKCD | 0.0194 | −0.0539 | 0.1518 | NS |
| PRDX1 | PRDX1 | 0.0266 | −0.0393 | 0.2556 | NS |
| Rab25 | RAB25 | 0.0432 | −0.0767 | 0.0011 | ** |
| Rad50 | RAD50 | 0.0579 | −0.0170 | 0.1381 | NS |
| Shc_pY317 | SHC1 | 0.0064 | −0.0834 | 0.0026 | ** |
| Src_pY416 | SRC | 0.0296 | −0.0103 | 0.4421 | NS |
| VEGFR2 | KDR | 0.0142 | −0.0164 | 0.3048 | NS |
CCNF, cyclin F; RPPA, reverse-phase protein array. ****, extremely significant (P<0.0001); ***, extremely significant (P=0.0001 to 0.001); **, very significant (P=0.001 to 0.01); *, significant (P=0.01 to 0.05); NS, not significant (P≥0.05).
Table V.
Expression of proteins which positively correlate with CCNF.
| CCNFdownregulated | CCNFupregulated | ||||
|---|---|---|---|---|---|
| RPPA (z-score) | |||||
| Protein | Gene | Downregulated | Upregulated | P-value | Significance |
| 4E-BP1 | EIF4EBP1 | −0.0327 | 0.0285 | 0.3383 | NS |
| 4E-BP1_pS65 | EIF4EBP1 | −0.0677 | 0.0891 | <0.0001 | **** |
| 4E-BP1_pT70 | EIF4EBP1 | −0.0655 | 0.0860 | <0.0001 | **** |
| ACC_pS79 | ACACA | −0.0054 | 0.0362 | 0.3200 | NS |
| C-Raf | RAF1 | −0.0110 | 0.0031 | 0.3485 | NS |
| CDK1_pY15 | CDK1 | −0.1318 | 0.1145 | <0.0001 | **** |
| Chk1 | CHEK1 | −0.0156 | 0.0301 | 0.0333 | * |
| Chk2 | CHEK2 | −0.0137 | 0.0578 | 0.0421 | * |
| Cyclin_B1 | CCNB1 | −0.2619 | 0.2546 | <0.0001 | **** |
| Cyclin_E1 | CCNE1 | −0.0766 | 0.1020 | 0.0009 | *** |
| eEF2 | EEF2 | −0.0851 | 0.0434 | 0.0449 | * |
| FoxM1 | FOXM1 | −0.0423 | 0.1525 | <0.0001 | **** |
| GAPDH | GAPDH | −0.0492 | 0.0336 | 0.5952 | NS |
| MIG-6 | ERRFI1 | −0.0025 | 0.0536 | 0.1141 | NS |
| MSH2 | MSH2 | −0.0296 | 0.0032 | 0.3485 | NS |
| MSH6 | MSH6 | −0.1614 | 0.0359 | 0.0003 | *** |
| NF-kB-p65_pS536 | NFKB1 | −0.0494 | 0.0028 | 0.6642 | NS |
| NF2 | NF2 | −0.0131 | 0.0148 | 0.7265 | NS |
| p21 | CDKN1A | −0.0769 | 0.0860 | 0.0186 | * |
| p38_MAPK | MAPK14 | −0.0104 | 0.0141 | 0.7464 | NS |
| p62-LCK-ligand | SQSTM1 | −0.0667 | 0.0042 | 0.1143 | NS |
| PARP1 | PARP1 | −0.0340 | 0.1803 | 0.0425 | * |
| PCNA | PCNA | −0.0654 | 0.1086 | <0.0001 | **** |
| PRAS40_pT246 | AKT1S1 | −0.0248 | 0.0148 | 0.1293 | NS |
| Rb_pS807_S811 | RB1 | −0.1385 | 0.1091 | 0.0014 | ** |
| S6_pS240_S244 | RPS6KB1 | −0.0474 | 0.1577 | 0.0086 | ** |
| SLC1A5 | SLC1A5 | −0.0542 | 0.0092 | 0.2129 | NS |
| Src | SRC | −0.0137 | 0.0101 | 0.4254 | NS |
| Src_pY527 | SRC | −0.0814 | 0.1241 | 0.0022 | ** |
| TFRC | TFRC | −0.1894 | 0.3049 | <0.0001 | **** |
| Tuberin_pT1462 | TSC2 | −0.0626 | 0.0410 | 0.0488 | * |
| XRCC1 | XRCC1 | −0.0784 | 0.0078 | 0.0065 | ** |
CCNF, cyclin F; RPPA, reverse-phase protein array. ****, extremely significant (P<0.0001); ***, extremely significant (P=0.0001 to 0.001); **, very significant (P=0.001 to 0.01); *, significant (P=0.01 to 0.05); NS, not significant (P≥0.05).
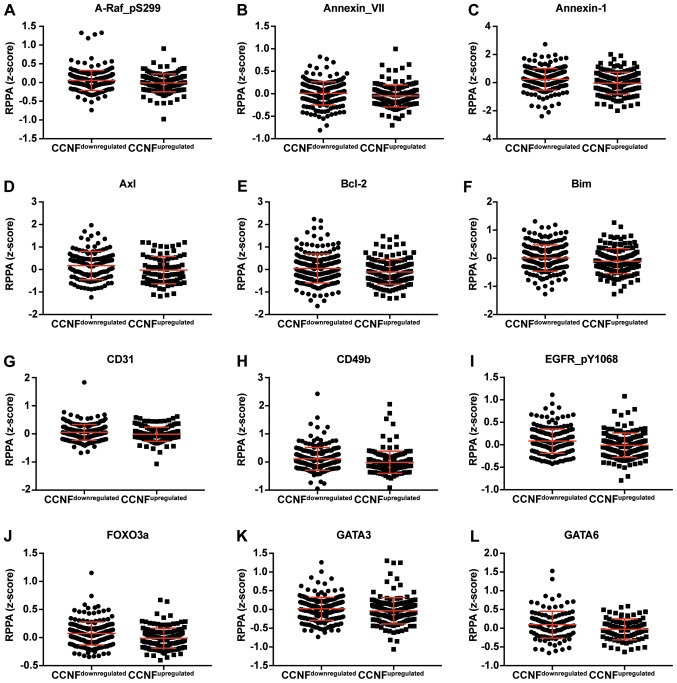
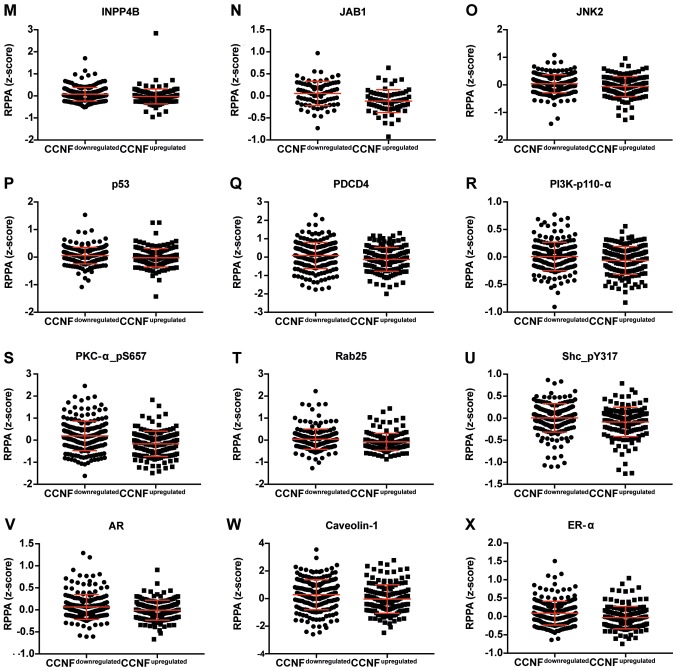
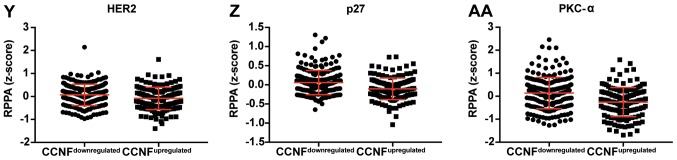
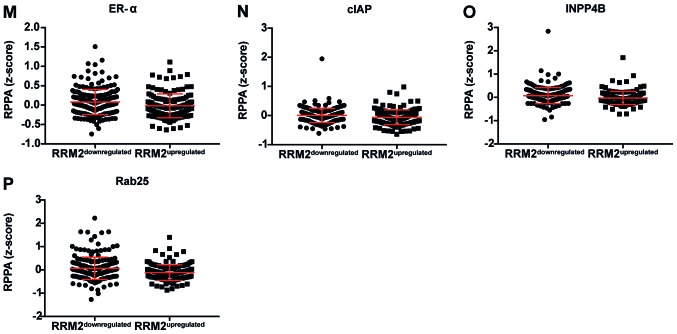
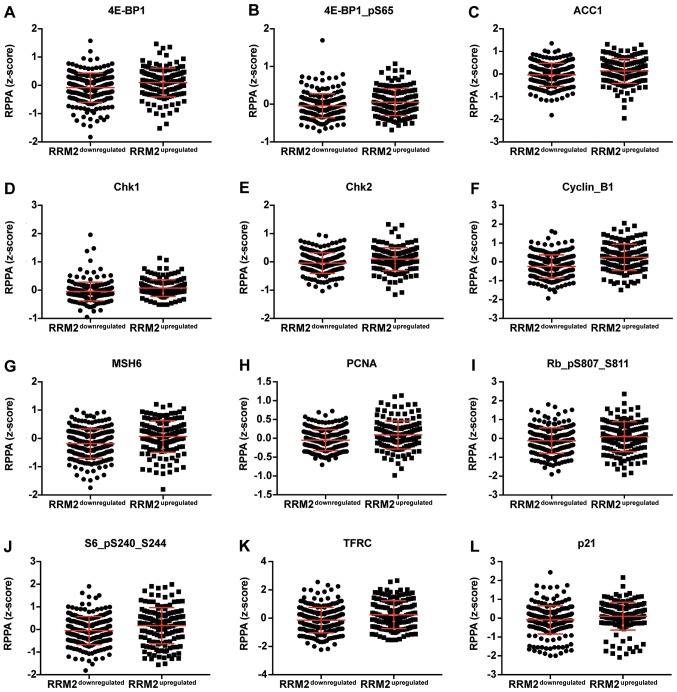
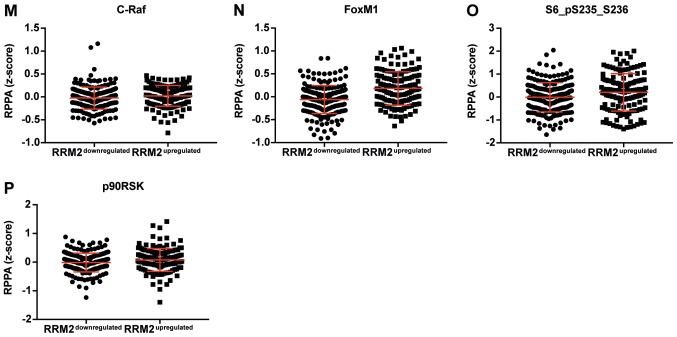
Figure 3.
Dot plot representation of the protein levels by RPPA (z-score). (A-L) Proteins negatively correlated with CCNF mRNA. Horizontal bars represent lower quartile, median and higher quartile. CCNF, cyclin F; RPPA, reverse-phase protein array. Dot plot representation of the protein levels by RPPA (z-score). (M-AA) Proteins negatively correlated with CCNF mRNA. Horizontal bars represent lower quartile, median and higher quartile. CCNF, cyclin F; RPPA, reverse-phase protein array.
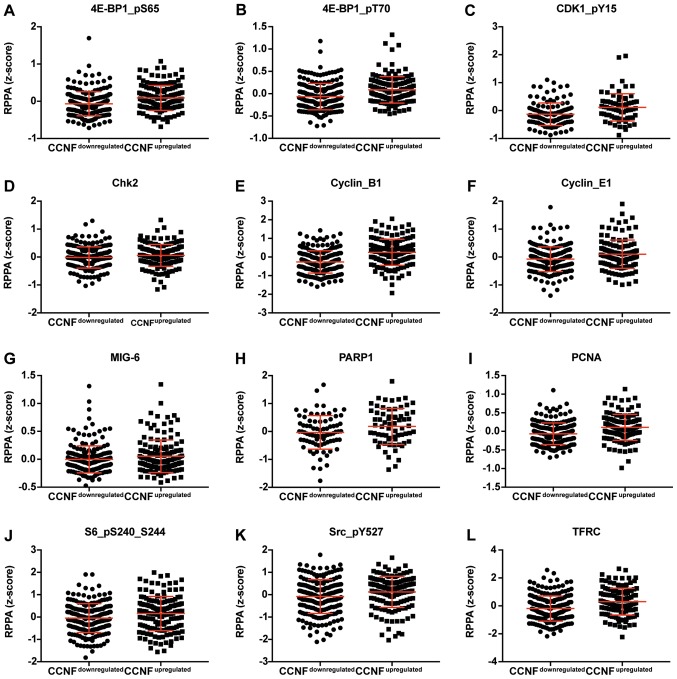
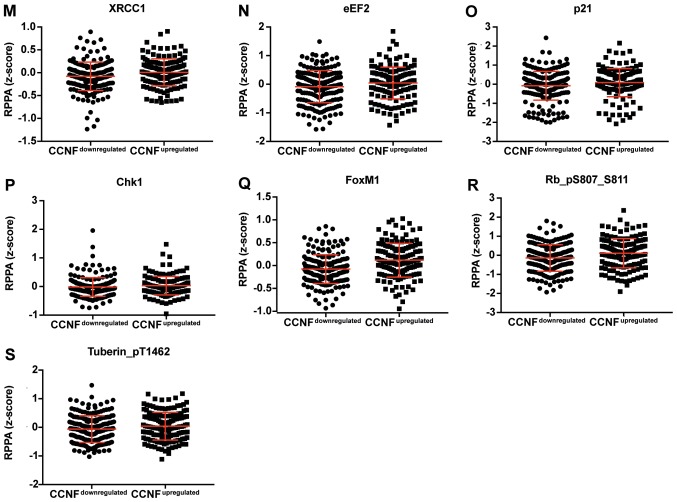
Figure 4.
Dot plot representation of the protein levels by RPPA (z-score). (A-S) Proteins positively correlated with CCNF mRNA. Horizontal bars represent lower quartile, median and higher quartile. CCNF, cyclin F; RPPA, reverse-phase protein array.
Table VII.
Expression of proteins which are negatively correlated with RRM2.
| RRM2downregulated | RRM2upregulated | ||||
|---|---|---|---|---|---|
| RPPA (z-score) | |||||
| Protein | Gene | Upregulated | Downregulated | P-value | Significance |
| 14-3-3_ζ | YWHAZ | 0.0436 | −0.0176 | 0.1293 | NS |
| α-catenin | CTNNB1 | 0.0676 | −0.0037 | 0.0957 | NS |
| AMPK_α | PRKAA1 | 0.0076 | −0.0467 | 0.0311 | * |
| Bcl-2 | BCL2 | 0.1080 | −0.1967 | <0.0001 | **** |
| cIAP | BIRC2 | 0.0042 | −0.0600 | 0.0043 | ** |
| E-cadherin | CDH1 | 0.1282 | −0.4331 | 0.0003 | *** |
| ER-α | ESR1 | 0.0796 | −0.0194 | 0.0013 | ** |
| FOXO3a | FOXO3 | 0.0664 | −0.0035 | 0.0041 | ** |
| GATA3 | GATA3 | 0.0059 | −0.0189 | 0.0886 | NS |
| HER2 | ERBB2 | 0.0414 | −0.0487 | 0.0298 | * |
| HER3 | ERBB3 | 0.0900 | −0.1863 | <0.0001 | **** |
| INPP4B | INPP4B | 0.0749 | −0.0261 | 0.0007 | *** |
| JAB1 | COPS5 | 0.0083 | −0.0772 | 0.0957 | NS |
| JNK2 | MAPK9 | 0.0047 | −0.0109 | 0.6772 | NS |
| p27_pT198 | CDKN1B | 0.0017 | −0.0069 | 0.7788 | NS |
| p38_MAPK | MAPK14 | 0.0349 | −0.0488 | 0.0165 | * |
| p38_pT180_Y182 | MAPK14 | 0.0086 | −0.0314 | 0.4440 | NS |
| PARP_cleaved | PARP1 | 0.0073 | −0.0035 | 0.4065 | NS |
| PDCD4 | PDCD4 | 0.1225 | −0.1818 | 0.0001 | *** |
| PDK1 | PDPK1 | 0.0294 | −0.0014 | 0.1348 | NS |
| PDK1_pS241 | PDPK1 | 0.0071 | −0.0442 | 0.2341 | NS |
| PI3K-p85 | PIK3R1 | 0.0116 | −0.0613 | 0.0605 | NS |
| PKC-α | PRKCA | 0.0155 | −0.0969 | 0.0414 | * |
| PKC-α_pS657 | PRKCA | 0.0906 | −0.0247 | 0.0307 | * |
| PRDX1 | PRDX1 | 0.0146 | −0.0238 | 0.5525 | NS |
| PREX1 | PREX1 | 0.0619 | −0.0082 | 0.2623 | NS |
| Rab25 | RAB25 | 0.0707 | −0.1177 | <0.0001 | **** |
| Rad50 | RAD50 | 0.0570 | −0.0174 | 0.0624 | NS |
| Src | SRC | 0.0459 | −0.0729 | 0.0033 | ** |
| Src_pY527 | SRC | 0.0315 | −0.0300 | 0.3020 | NS |
| VEGFR2 | KDR | 0.0191 | −0.0239 | 0.4077 | NS |
| YAP | YAP1 | 0.0412 | −0.0283 | 0.0215 | * |
| YAP_pS127 | YAP1 | 0.1242 | −0.0491 | 0.0106 | * |
RRM2, ribonucleotide reductase family member 2; RPPA, reverse-phase protein array. ****, extremely significant (P<0.0001); ***, extremely significant (P=0.0001 to 0.001); **, very significant (P=0.001 to 0.01); *, significant (P=0.01 to 0.05); NS, not significant (P≥0.05).
Table IX.
Expression of proteins which are positively correlated with RRM2.
| RRM2downregulated | RRM2upregulated | ||||
|---|---|---|---|---|---|
| RPPA (z-score) | |||||
| Protein | Gene | Downregulated | Upregulated | P-value | Significance |
| 4E-BP1 | EIF4EBP1 | −0.0824 | 0.0995 | 0.0010 | *** |
| 4E-BP1_pS65 | EIF4EBP1 | −0.0413 | 0.0554 | 0.0126 | * |
| 4E-BP1_pT70 | EIF4EBP1 | −0.0152 | 0.0185 | 0.2779 | NS |
| ACC_pS79 | ACACA | −0.0209 | 0.0587 | 0.0782 | NS |
| ACC1 | ACACA | −0.0340 | 0.1352 | 0.0024 | ** |
| Bax | BAX | −0.0251 | 0.0043 | 0.8262 | NS |
| C-Raf | RAF1 | −0.0244 | 0.0222 | 0.0089 | ** |
| CDK1_pY15 | CDK1 | −0.0711 | 0.0147 | 0.1638 | NS |
| Chk1 | CHEK1 | −0.0460 | 0.0737 | <0.0001 | **** |
| Chk1_pS345 | CHEK1 | −0.0142 | 0.0205 | 0.0618 | NS |
| Chk2 | CHEK2 | −0.0412 | 0.0977 | 0.0002 | *** |
| Cyclin_B1 | CCNB1 | −0.2434 | 0.2391 | <0.0001 | **** |
| Cyclin_E1 | CCNE1 | −0.0330 | 0.0445 | 0.1473 | NS |
| eEF2 | EEF2 | −0.0765 | 0.0339 | 0.1272 | NS |
| EGFR_pY1173 | EGFR | −0.0088 | 0.0283 | 0.1524 | NS |
| eIF4E | EIF4E | −0.0487 | 0.0033 | 0.1951 | NS |
| FoxM1 | FOXM1 | −0.0608 | 0.1823 | <0.0001 | **** |
| GAPDH | GAPDH | −0.0538 | 0.0416 | 0.0996 | NS |
| HER3_pY1289 | ERBB3 | −0.0062 | 0.0066 | 0.3676 | NS |
| MSH2 | MSH2 | −0.0493 | 0.0314 | 0.0703 | NS |
| MSH6 | MSH6 | −0.1812 | 0.0677 | <0.0001 | **** |
| Myosin-IIa | MYH9 | −0.0317 | 0.0371 | 0.4099 | NS |
| NF2 | NF2 | −0.0205 | 0.0258 | 0.2552 | NS |
| p21 | CDKN1A | −0.0880 | 0.1049 | 0.0025 | ** |
| p62-LCK-ligand | SQSTM1 | −0.0850 | 0.0313 | 0.1070 | NS |
| p90RSK | RPS6KA1 | −0.0133 | 0.0750 | 0.0363 | * |
| PCNA | PCNA | −0.0499 | 0.0905 | 0.0001 | *** |
| PRAS40_pT246 | AKT1S1 | −0.0225 | 0.0124 | 0.3327 | NS |
| Rb_pS807_S811 | RB1 | −0.1222 | 0.0913 | 0.0035 | ** |
| S6_pS235_S236 | RPS6KB1 | −0.0110 | 0.2044 | 0.0053 | ** |
| S6_pS240_S244 | RPS6KB1 | −0.0516 | 0.1676 | 0.0024 | ** |
| SLC1A5 | SLC1A5 | −0.0858 | 0.0421 | 0.0743 | NS |
| Src_pY416 | SRC | −0.0307 | 0.0735 | 0.0630 | NS |
| TFRC | TFRC | −0.1404 | 0.2463 | 0.0007 | *** |
| Transglutaminase | TGM1 | −0.0275 | 0.0094 | 0.5674 | NS |
| TSC1 | TSC1 | −0.0611 | 0.0051 | 0.1200 | NS |
RRM2, ribonucleotide reductase family member 2; RPPA, reverse-phase protein array. ****, extremely significant (P<0.0001); ***, extremely significant (P=0.0001 to 0.001); **, very significant (P=0.001 to 0.01); *, significant (P=0.01 to 0.05); NS, not significant (P≥0.05).
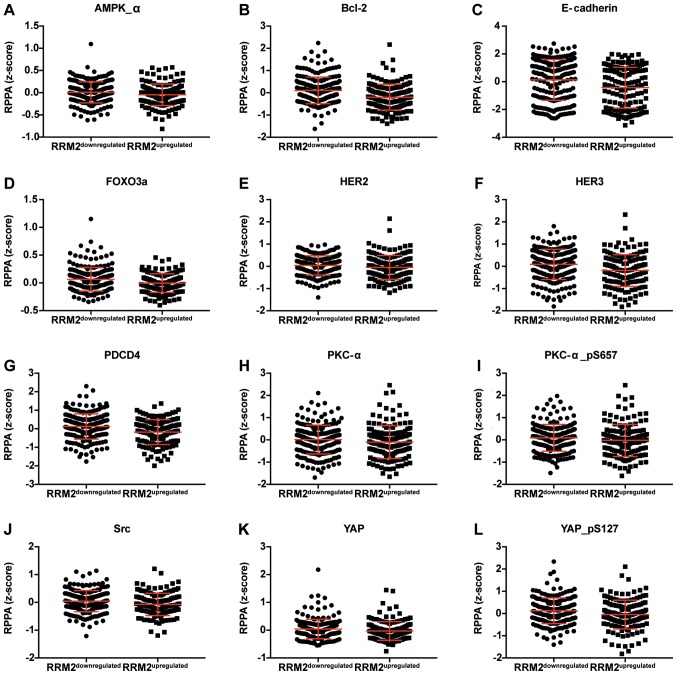
Figure 5.
Dot plot representation of the protein levels by RPPA (z-score). (A-P) Proteins negatively correlated with RRM2 mRNA. Horizontal bars represent lower quartile, median and higher quartile. RRM2, ribonucleotide reductase family member 2; RPPA, reverse-phase protein array.
Figure 6.
Dot plot representation of the protein levels by RPPA (z-score). (A-P) Proteins positively correlated with RRM2 mRNA. Horizontal bars represent lower quartile, median and higher quartile. RRM2, ribonucleotide reductase family member 2; RPPA, reverse-phase protein array.
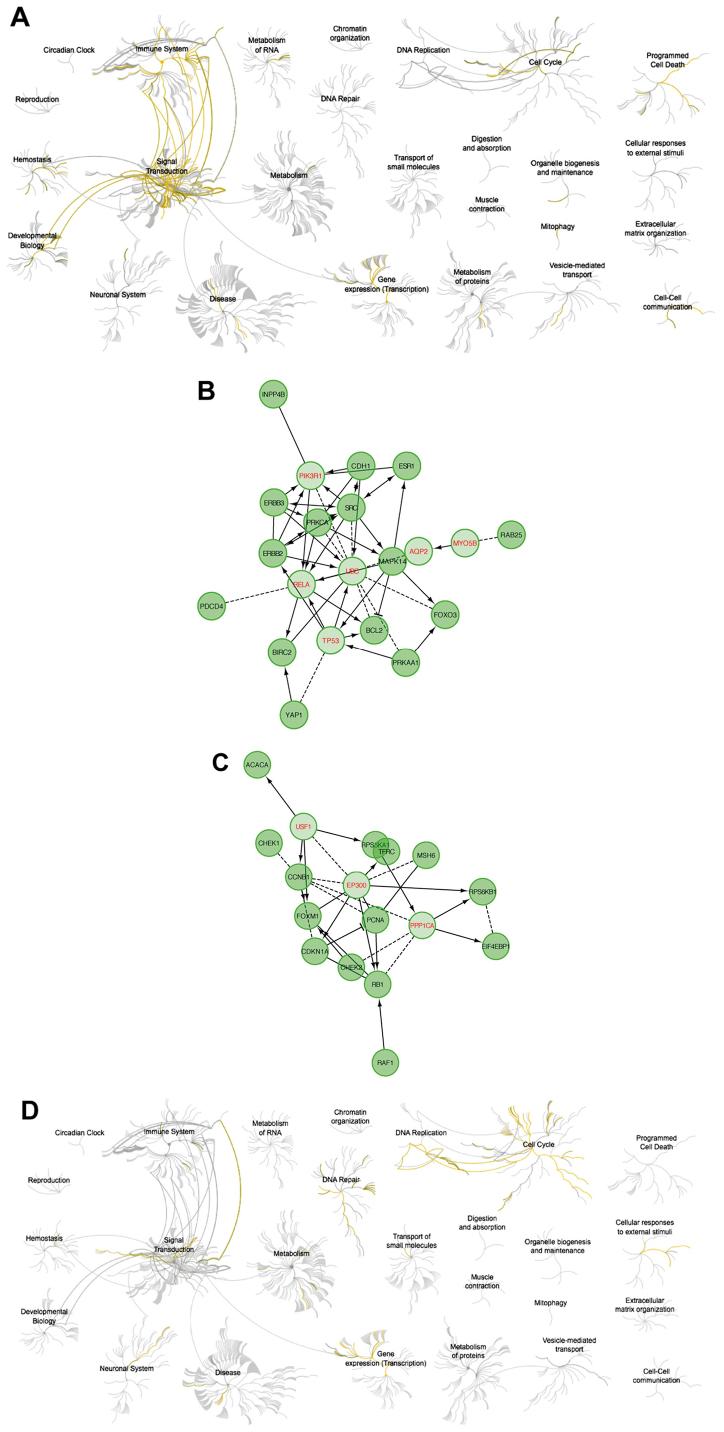
The analysis using Reactome showed that upregulation of cyclin F resulted in downregulation of pathways responsible for signal transduction and activation of cell cycle-related and DNA repair (Fig. 7). High expression of RRM2 mRNA also resulted in downregulation of cell signaling pathways. Activation of the cell cycle and DNA pathways was also visible but less univocal (Fig. 8). Upregulation of cyclin F coincides with altered expression of factors that were associated with worse patient outcome. Furthermore, patients with worse outcome had increased levels of proliferative proteins, such as cyclin E, cyclin B, PCNA, pro-survival factors such as p27 or FOXM1 and connected with AKT pathway activation (INPP4B). The list of biological processes altered by cyclin F dysregulation are presented in Tables IV and VI. Furthermore, data presenting biological processes influenced by changes in RRM2 expression are presented in Tables VIII and X.
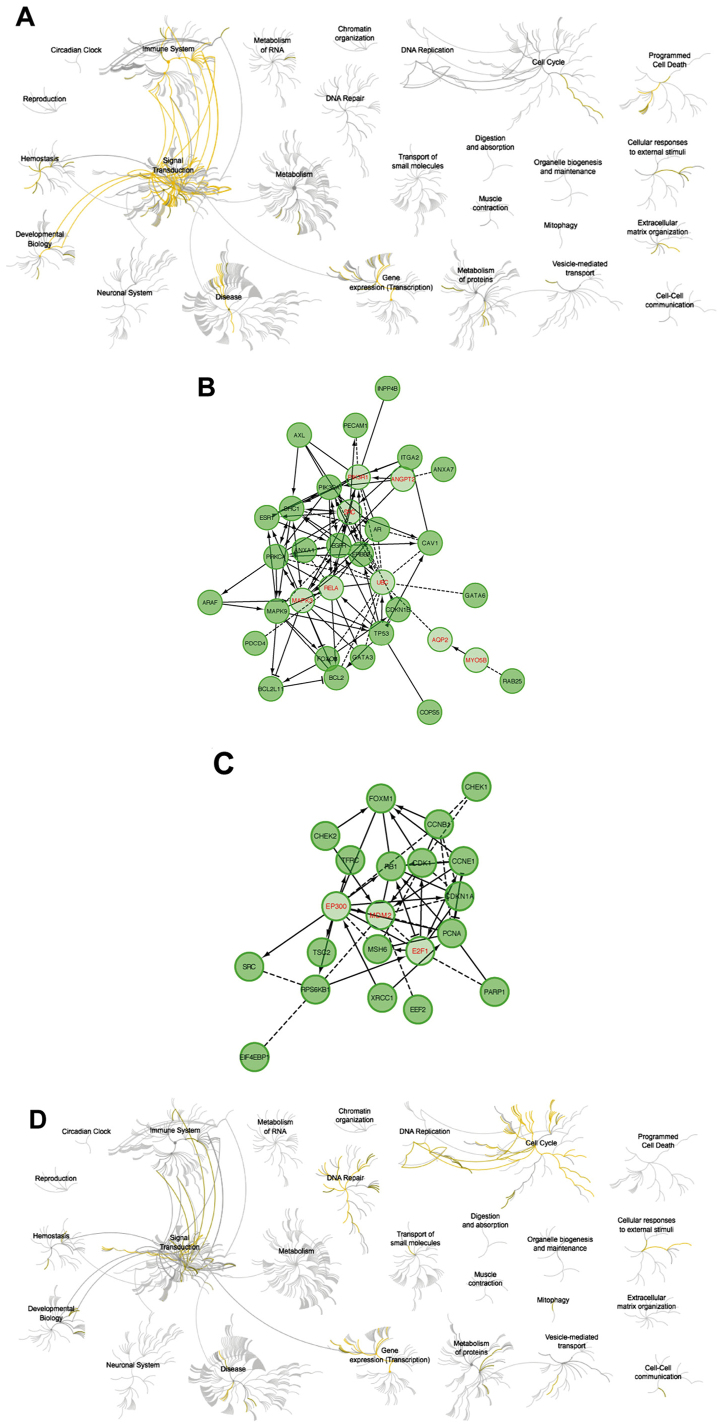
Figure 7.
(A) Pathways negatively correlated with CCNF expression. (B) Relationships between proteins negatively correlated with CCNF expression involved in pathway analysis. (C) Pathways positively correlated with CCNF expression. (D) Relationships between proteins positively correlated with CCNF expression. CCNF, cyclin F.
Figure 8.
(A) Pathways negatively correlated with RRM2 expression. (B) Relationships between proteins negatively correlated with RRM2 expression involved in pathway analysis. (C) Pathways positively correlated with RRM2 expression. (D) Relationships between proteins positively correlated with RRM2 expression. RRM2, ribonucleotide reductase family member 2.
Table IV.
Biological process and pathway analysis of genes whose products are negatively correlated with CCNF expression.
| Factor | P-value | Number of genes | Gene list |
|---|---|---|---|
| Biological process | |||
| Regulation of apoptotic process | 1.26E-12 | 19 | GATA3, GATA6, CDKN1B, FOXO3, PRKCA, ERBB2, BCL2, CAV1, MAPK9, BCL2L11, EGFR, PIK3CA, ANXA1, AXL, AR, ARAF, PDCD4, ESR1, TP53 |
| Regulation of intracellular signal transduction | 7.99E-12 | 19 | SHC1, GATA3, PRKCA, ERBB2, BCL2, CAV1, MAPK9, BCL2L11, EGFR, PIK3CA, COPS5, AXL, AR, ARAF, PDCD4, ESR1, INPP4B, TP53, PECAM1 |
| Apoptotic process | 2.63E-11 | 19 | GATA3, GATA6, CDKN1B, FOXO3, PRKCA, ERBB2, BCL2, CAV1, MAPK9, BCL2L11, EGFR, PIK3CA, ANXA1, AXL, AR, ARAF, PDCD4, ESR1, TP53 |
| Negative regulation of apoptotic process | 3.01E-11 | 15 | GATA3, GATA6, CDKN1B, PRKCA, ERBB2, BCL2, CAV1, EGFR, PIK3CA, ANXA1, AXL, AR, ARAF, PDCD4, TP53 |
| Positive regulation of cellular protein metabolic process | 1.08E-10 | 17 | SHC1, GATA3, CDKN1B, PRKCA, ERBB2, BCL2, ITGA2, CAV1, MAPK9, BCL2L11, EGFR, PIK3CA, AR, ARAF, ESR1, TP53, PECAM1 |
| Regulation of protein modification process | 1.30E-10 | 18 | SHC1, GATA3, CDKN1B, PRKCA, ERBB2, BCL2, ITGA2, CAV1, MAPK9, EGFR, PIK3CA, COPS5, AR, ARAF, PDCD4, ESR1, TP53, PECAM1 |
| Positive regulation of signaling | 3.37E-10 | 17 | SHC1, GATA3, GATA6, PRKCA, ERBB2, BCL2, ITGA2, CAV1, MAPK9, BCL2L11, EGFR, AXL, AR, ARAF, ESR1, TP53, PECAM1 |
| Positive regulation of cell communication | 3.61E-10 | 17 | SHC1, GATA3, GATA6, PRKCA, ERBB2, BCL2, ITGA2, CAV1, MAPK9, BCL2L11, EGFR, AXL, AR, ARAF, ESR1, TP53, PECAM1 |
| Regulation of phosphorylation | 7.58E-10 | 16 | SHC1, CDKN1B, PRKCA, ERBB2, BCL2, CAV1, MAPK9, EGFR, PIK3CA, COPS5, AR, ARAF, PDCD4, ESR1, TP53, PECAM1 |
| Positive regulation of phosphorylation | 7.76E-10 | 14 | SHC1, CDKN1B, PRKCA, ERBB2, BCL2, CAV1, MAPK9, EGFR, PIK3CA, AR, ARAF, ESR1, TP53, PECAM1 |
| Regulation of cell proliferation | 2.67E-09 | 16 | SHC1, GATA3, GATA6, CDKN1B, FOXO3, PRKCA, ERBB2, BCL2, RAB25, ITGA2, CAV1, EGFR, ANXA1, AR, ESR1, TP53 |
| Positive regulation of cell proliferation | 4.30E-09 | 13 | SHC1, GATA6, CDKN1B, PRKCA, ERBB2, BCL2, RAB25, ITGA2, CAV1, EGFR, ANXA1, AR, ESR1 |
| Cell adhesion | 1.11E-07 | 14 | SHC1, GATA3, PRKCA, ERBB2, BCL2, ITGA2, CAV1, BCL2L11, EGFR, PIK3CA, ANXA1, AXL, TP53, PECAM1 |
| Positive regulation of apoptotic process | 3.01E-07 | 10 | GATA6, CDKN1B, FOXO3, BCL2, CAV1, MAPK9, BCL2L11, ANXA1, PDCD4, TP53 |
| Pathway | |||
| EGFR tyrosine kinase inhibitor resistance | 2.50E-13 | 10 | SHC1, FOXO3, PRKCA, ERBB2, BCL2, BCL2L11, EGFR, PIK3CA, AXL, ARAF |
| Endocrine resistance | 9.60E-13 | 10 | SHC1, CDKN1B, ERBB2, BCL2, MAPK9, EGFR, PIK3CA, ARAF, ESR1, TP53 |
| Proteoglycans in cancer | 1.01E-09 | 10 | PRKCA, ERBB2, ITGA2, CAV1, EGFR, PIK3CA, ARAF, PDCD4, ESR1, TP53 |
| ErbB signaling pathway | 1.01E-09 | 8 | SHC1, CDKN1B, PRKCA, ERBB2, MAPK9, EGFR, PIK3CA, ARAF |
| Focal adhesion | 1.57E-08 | 9 | SHC1, PRKCA, ERBB2, BCL2, ITGA2, CAV1, MAPK9, EGFR, PIK3CA |
| Pathways in cancer | 1.57E-08 | 11 | CDKN1B, PRKCA, ERBB2, BCL2, ITGA2, MAPK9, EGFR, PIK3CA, AR, ARAF, TP53 |
| MicroRNAs in cancer | 2.01E-08 | 10 | SHC1, CDKN1B, PRKCA, ERBB2, BCL2, BCL2L11, EGFR, PIK3CA, PDCD4, TP53 |
| FoxO signaling pathway | 4.15E-07 | 7 | CDKN1B, FOXO3, MAPK9, BCL2L11, EGFR, PIK3CA, ARAF |
| Signaling by SCF-KIT | 7.61E-07 | 9 | SHC1, CDKN1B, FOXO3, PRKCA, ERBB2, EGFR, PIK3CA, ARAF, TP53 |
| PI3K-Akt signaling pathway | 7.61E-07 | 9 | CDKN1B, FOXO3, PRKCA, BCL2, ITGA2, BCL2L11, EGFR, PIK3CA, TP53 |
| Signaling by NGF | 9.31E-07 | 10 | SHC1, CDKN1B, FOXO3, PRKCA, ERBB2, BCL2L11, EGFR, PIK3CA, ARAF, TP53 |
| HIF-1 signaling pathway | 1.43E-06 | 6 | CDKN1B, PRKCA, ERBB2, BCL2, EGFR, PIK3CA |
| Apoptosis signaling pathway | 1.43E-06 | 6 | PRKCA, BCL2, MAPK9, BCL2L11, PIK3CA, TP53 |
| Signaling by ERBB2 | 1.54E-06 | 5 | SHC1, PRKCA, ERBB2, EGFR, PIK3CA |
CCNF, cyclin F; SCF, Skp, Cullin, F-box containing. P-values were corrected for multiple comparisons using the false discovery rate (FDR) (Benjamini and Hochberg).
Table VI.
Biological process and pathway analysis of genes whose products are positively correlated with CCNF expression.
| Factor | P-value | Number of genes | Gene list |
|---|---|---|---|
| Biological process | |||
| Regulation of cell cycle | 1.88E-10 | 13 | CHEK2, FOXM1, CCNE1, CDKN1A, RB1, TSC2, RPS6KB1, CDK1, CHEK1, PCNA, EIF4EBP1, SRC, CCNB1 |
| Cell cycle phase transition | 1.94E-10 | 11 | CHEK2, FOXM1, CCNE1, CDKN1A, RB1, RPS6KB1, CDK1, CHEK1, PCNA, EIF4EBP1, CCNB1 |
| Cell cycle G1/S phase transition | 2.22E-10 | 9 | CHEK2, CCNE1, CDKN1A, RB1, RPS6KB1, CDK1, PCNA, EIF4EBP1, CCNB1 |
| Positive regulation of cell cycle | 1.82E-09 | 9 | CHEK2, CDKN1A, RB1, RPS6KB1, CDK1, PCNA, EIF4EBP1, SRC, CCNB1 |
| Regulation of DNA metabolic process | 2.32E-09 | 9 | CHEK2, FOXM1, CDKN1A, MSH6, PARP1, CDK1, CHEK1, PCNA, SRC |
| Cell cycle arrest | 4.10E-09 | 8 | CHEK2, FOXM1, CDKN1A, RB1, TSC2, CDK1, PCNA, CCNB1 |
| Negative regulation of mitotic cell cycle phase transition | 8.06E-09 | 7 | CHEK2, CDKN1A, RB1, CDK1, CHEK1, PCNA, CCNB1 |
| Negative regulation of G1/S transition of mitotic cell cycle | 2.07E-08 | 6 | CHEK2, CDKN1A, RB1, CDK1, PCNA, CCNB1 |
| DNA damage checkpoint | 1.70E-07 | 6 | CHEK2, CDKN1A, CDK1, CHEK1, PCNA, CCNB1 |
| Positive regulation of macromolecule biosynthetic process | 1.94E-07 | 12 | CHEK2, FOXM1, CCNE1, RB1, PARP1, TSC2, EEF2, RPS6KB1, CDK1, CHEK1, PCNA, SRC |
| DNA repair | 3.08E-07 | 8 | CHEK2, FOXM1, MSH6, PARP1, CDK1, CHEK1, PCNA, XRCC1 |
| Positive regulation of gene expression | 3.21E-07 | 12 | CHEK2, FOXM1, CCNE1, RB1, PARP1, TSC2, EEF2, RPS6KB1, CDK1, CHEK1, SRC, CCNB1 |
| Positive regulation of cell cycle arrest | 3.74E-07 | 5 | CHEK2, CDKN1A, CDK1, PCNA, CCNB1 |
| Positive regulation of cellular biosynthetic process | 3.74E-07 | 12 | CHEK2, FOXM1, CCNE1, RB1, PARP1, TSC2, EEF2, RPS6KB1, CDK1, CHEK1, PCNA, SRC |
| Pathway | |||
| Cell cycle | 1.06E-09 | 8 | CHEK2, CCNE1, CDKN1A, RB1, CDK1, CHEK1, PCNA, CCNB1 |
| p53 signaling pathway | 1.06E-09 | 7 | CHEK2, CCNE1, CDKN1A, TSC2, CDK1, CHEK1, CCNB1 |
| FOXM1 transcription factor network | 1.61E-09 | 6 | CHEK2, FOXM1, RB1, CDK1, CCNB1, XRCC1 |
| E2F mediated regulation of DNA replication | 9.24E-08 | 5 | CCNE1, RB1, CDK1, PCNA, CCNB1 |
| mTOR signaling pathway | 9.46E-07 | 5 | CCNE1, TSC2, EEF2, RPS6KB1, EIF4EBP1 |
| ATM signaling pathway | 4.84E-05 | 3 | CHEK2, CDKN1A, CHEK1 |
| DNA double-strand break repair | 5.32E-05 | 5 | CHEK2, PARP1, CHEK1, PCNA, XRCC1 |
| ErbB signaling pathway | 8.14E-05 | 4 | CDKN1A, RPS6KB1, EIF4EBP1, SRC |
| Endocrine resistance | 1.16E-04 | 4 | CDKN1A, RB1, RPS6KB1, SRC |
| HIF-1 signaling pathway | 1.39E-04 | 4 | CDKN1A, RPS6KB1, EIF4EBP1, TFRC |
| Base excision repair | 1.57E-04 | 3 | PARP1, PCNA, XRCC1 |
| AMPK signaling pathway | 2.39E-04 | 4 | TSC2, EEF2, RPS6KB1, EIF4EBP1 |
| PI3K-Akt signaling pathway | 7.78E-04 | 5 | CCNE1, CDKN1A, TSC2, RPS6KB1, EIF4EBP1 |
| Mismatch repair | 1.42E-03 | 2 | MSH6, PCNA |
CCNF, cyclin F. P-values were corrected for multiple comparisons using the false discovery rate (FDR) (Benjamini and Hochberg).
Table VIII.
Biological process and pathway analysis of genes whose products are negatively correlated with RRM2 expression.
| Factor | P-value | Number of genes | Gene list |
|---|---|---|---|
| Biological process | |||
| Regulation of cell proliferation | 4.03E-09 | 13 | FOXO3, CDH1, BIRC2, PRKCA, YAP1, ERBB2, ERBB3, ESR1, BCL2, RAB25, MAPK14, PRKAA1, SRC |
| Apoptotic process | 1.03E-08 | 13 | FOXO3, CDH1, BIRC2, PRKCA, YAP1, ERBB2, ERBB3, PDCD4, ESR1, BCL2, MAPK14, PRKAA1, SRC |
| Regulation of apoptotic process | 1.39E-08 | 12 | FOXO3, CDH1, BIRC2, PRKCA, YAP1, ERBB2, ERBB3, PDCD4, ESR1, BCL2, PRKAA1, SRC |
| Negative regulation of signal transduction | 2.30E-08 | 11 | FOXO3, CDH1, PRKCA, YAP1, ERBB3, PDCD4, ESR1, BCL2, MAPK14, PRKAA1, SRC |
| Negative regulation of apoptotic process | 8.07E-07 | 9 | BIRC2, PRKCA, YAP1, ERBB2, ERBB3, PDCD4, BCL2, PRKAA1, SRC |
| Regulation of intracellular signal transduction | 8.07E-07 | 11 | BIRC2, PRKCA, ERBB2, ERBB3, PDCD4, ESR1, BCL2, INPP4B, MAPK14, PRKAA1, SRC |
| Positive regulation of intracellular signal transduction | 1.09E-06 | 9 | BIRC2, PRKCA, ERBB2, ERBB3, ESR1, BCL2, MAPK14, PRKAA1, SRC |
| Regulation of cell motility | 4.00E-06 | 8 | CDH1, PRKCA, ERBB2, ERBB3, BCL2, RAB25, MAPK14, SRC |
| Positive regulation of protein modification process | 5.45E-06 | 9 | BIRC2, PRKCA, ERBB2, ERBB3, ESR1, BCL2, MAPK14, PRKAA1, SRC |
| Regulation of cellular component movement | 6.29E-06 | 8 | CDH1, PRKCA, ERBB2, ERBB3, BCL2, RAB25, MAPK14, SRC |
| MAPK cascade | 8.78E-06 | 8 | PRKCA, ERBB2, ERBB3, PDCD4, ESR1, MAPK14, PRKAA1, SRC |
| Positive regulation of protein phosphorylation | 1.03E-05 | 8 | PRKCA, ERBB2, ERBB3, ESR1, BCL2, MAPK14, PRKAA1, SRC |
| Signal transduction by protein phosphorylation | 1.03E-05 | 8 | PRKCA, ERBB2, ERBB3, PDCD4, ESR1, MAPK14, PRKAA1, SRC |
| Regulation of canonical Wnt signaling pathway | 3.05E-05 | 5 | FOXO3, CDH1, YAP1, MAPK14, SRC |
| Pathway | |||
| EGFR tyrosine kinase inhibitor resistance | 1.74E-07 | 6 | FOXO3, PRKCA, ERBB2, ERBB3, BCL2, SRC |
| Proteoglycans in cancer | 5.39E-07 | 7 | PRKCA, ERBB2, ERBB3, PDCD4, ESR1, MAPK14, SRC |
| a6b1 and a6b4 Integrin signaling | 9.50E-06 | 4 | CDH1, PRKCA, ERBB2, ERBB3 |
| Endocrine resistance | 1.17E-05 | 5 | ERBB2, ESR1, BCL2, MAPK14, SRC |
| Signaling by ERBB2 | 4.47E-05 | 4 | PRKCA, ERBB2, ERBB3, SRC |
| Focal adhesion | 2.06E-04 | 5 | BIRC2, PRKCA, ERBB2, BCL2, SRC |
| ErbB signaling pathway | 2.06E-04 | 4 | PRKCA, ERBB2, ERBB3, SRC |
| NGF signalling via TRKA from the plasma membrane | 2.42E-04 | 6 | FOXO3, PRKCA, ERBB2, ERBB3, MAPK14, SRC |
| FAS (CD95) signaling pathway | 4.38E-04 | 3 | BIRC2, MAPK14, SRC |
| Signalling by NGF | 4.82E-04 | 6 | FOXO3, PRKCA, ERBB2, ERBB3, MAPK14, SRC |
| PI3K/AKT activation | 4.82E-04 | 4 | FOXO3, ERBB2, ERBB3, SRC |
| Cadherin signaling pathway | 6.77E-04 | 4 | CDH1, ERBB2, ERBB3, SRC |
| Pathways in cancer | 1.04E-03 | 5 | CDH1, BIRC2, PRKCA, ERBB2, BCL2 |
| Signaling by SCF-KIT | 6.93E-04 | 5 | FOXO3, PRKCA, ERBB2, ERBB3, SRC |
RRM2, ribonucleotide reductase family member 2. P-values were corrected for multiple comparisons using the false discovery rate (FDR) (Benjamini and Hochberg).
Table X.
Biological process and pathway analysis of genes whose products are positively correlated with RRM2 expression.
| Factor | P-value | Number of genes | Gene list |
|---|---|---|---|
| Biological process | |||
| Cell cycle phase transition | 1.99E-08 | 9 | CHEK2, FOXM1, CDKN1A, RB1, RPS6KB1, CHEK1, PCNA, EIF4EBP1, CCNB1 |
| Cell cycle G1/S phase transition | 9.08E-08 | 7 | CHEK2, CDKN1A, RB1, RPS6KB1, PCNA, EIF4EBP1, CCNB1 |
| Negative regulation of cell cycle phase transition | 1.97E-07 | 6 | CHEK2, CDKN1A, RB1, CHEK1, PCNA, CCNB1 |
| Cell cycle | 1.97E-07 | 11 | CHEK2, FOXM1, CDKN1A, RB1, MSH6, RPS6KA1, RPS6KB1, CHEK1, PCNA, EIF4EBP1, CCNB1 |
| Positive regulation of cell cycle | 2.96E-07 | 7 | CHEK2, CDKN1A, RB1, RPS6KB1, PCNA, EIF4EBP1, CCNB1 |
| Cell cycle process | 3.83E-07 | 10 | CHEK2, FOXM1, CDKN1A, RB1, MSH6, RPS6KB1, CHEK1, PCNA, EIF4EBP1, CCNB1 |
| Regulation of cell cycle | 5.63E-07 | 9 | CHEK2, FOXM1, CDKN1A, RB1, RPS6KB1, CHEK1, PCNA, EIF4EBP1, CCNB1 |
| Negative regulation of cell cycle G1/S phase transition | 5.94E-07 | 5 | CHEK2, CDKN1A, RB1, PCNA, CCNB1 |
| Regulation of cell cycle arrest | 7.05E-07 | 5 | CHEK2, FOXM1, CDKN1A, PCNA, CCNB1 |
| Signal transduction by p53 class mediator | 1.11E-06 | 6 | CHEK2, FOXM1, CDKN1A, CHEK1, PCNA, CCNB1 |
| Signal transduction in response to DNA damage | 1.11E-06 | 5 | CHEK2, FOXM1, CDKN1A, PCNA, CCNB1 |
| DNA integrity checkpoint | 3.58E-06 | 5 | CHEK2, CDKN1A, CHEK1, PCNA, CCNB1 |
| Regulation of cell proliferation | 1.36E-04 | 8 | FOXM1, CDKN1A, RB1, RAF1, RPS6KB1, CHEK1, CCNB1, TFRC |
| Regulation of cell growth | 2.26E-04 | 5 | FOXM1, CDKN1A, RB1, RPS6KA1, TFRC |
| Pathway | |||
| Cell cycle | 6.87E-07 | 6 | CHEK2, CDKN1A, RB1, CHEK1, PCNA, CCNB1 |
| Insulin signalling | 9.14E-06 | 4 | RAF1, RPS6KA1, RPS6KB1, EIF4EBP1 |
| FOXM1 transcription factor network | 9.14E-06 | 4 | CHEK2, FOXM1, RB1, CCNB1 |
| mTOR signaling pathway | 5.69E-05 | 4 | RAF1, RPS6KA1, RPS6KB1, EIF4EBP1 |
| p53 signaling pathway | 6.70E-05 | 4 | CHEK2, CDKN1A, CHEK1, CCNB1 |
| ATM signaling pathway | 9.01E-05 | 3 | CHEK2, CDKN1A, CHEK1 |
| ErbB signaling pathway | 1.04E-04 | 4 | CDKN1A, RAF1, RPS6KB1, EIF4EBP1 |
| HIF-1 signaling pathway | 1.53E-04 | 4 | CDKN1A, RPS6KB1, EIF4EBP1, TFRC |
| E2F mediated regulation of DNA replication | 2.21E-04 | 3 | RB1, PCNA, CCNB1 |
| G2/M DNA damage checkpoint | 2.21E-04 | 2 | CHEK1, CCNB1 |
| G1/S Transition | 2.34E-04 | 4 | CDKN1A, RB1, PCNA, CCNB1 |
| EGFR tyrosine kinase inhibitor resistance | 1.20E-03 | 3 | RAF1, RPS6KB1, EIF4EBP1 |
| RB tumor suppressor/checkpoint signaling in response to DNA damage | 1.25E-03 | 2 | RB1, CHEK1 |
| MAPKinase signaling pathway | 1.49E-03 | 3 | RAF1, RPS6KA1, RPS6KB1 |
RRM2, ribonucleotide reductase family member 2. P-values were corrected for multiple comparisons using the false discovery rate (FDR (Benjamini and Hochberg).
Discussion
There is only limited data describing cyclin F and its possible role in human cancer. D'Angiolella et al characterized the functional axis which is responsible for DNA repair following genotoxic stress (5). It is possible that interaction between cyclin F and RRM2 is significantly responsible for treatment response, thus detailed recognition of its nature may be useful for cancer clinical outcome prediction. Nuclear accumulation of RRM2, which allows efficient DNA repair, is preceded by downregulation of cyclin F. As it has been shown by D'Angiolella et al the insertion of wild-type cyclin F into hTERT RPE-1 cells prevents transposition of RRM2 from the cytoplasm to the nucleus (5). It has also been shown that overexpression of RRM2 may affect the proliferation of melanoma cells, their response to treatment in vivo, and is associated with worse overall survival in melanoma patients bearing mutations in the BRAF oncogene (8,11,12). Based on these data, we hypothesized that low expression of cyclin F in melanoma patients can be related to a poorer prognosis. This hypothesis was strengthened by the fact that the relationship between low cyclin F expression and poorer prognosis was demonstrated by Fu et al in patients with hepatocellular carcinoma. They showed that downregulation of cyclin F in hepatocellular carcinoma tissue samples was related to larger tumor size and poor tumor differentiation (13). Interestingly our analysis revealed that high expression of cyclin F mRNA is associated with poorer prognosis in skin cutaneous melanoma. Much as the result differs from what was expected, it is not surprising as overexpression of cyclin proteins is more common in cancer rather than their downregulation. Sun et al showed that overexpression of cyclin B1 is associated with poorer prognosis and reduced overall survival in breast cancer (14). Li et al revealed an association between high expression of cyclin B1 and claudin-1 with worse outcome in patients with hypopharyngeal squamous cell carcinoma (15). On the other hand, high cyclin B1 expression was found to reduce lymph node metastasis and distant metastasis stage, and was also associated with higher survival rates in colorectal cancer (16). High expression of cyclin D1 is a poor prognostic factor in gastric, oropharyngeal and breast cancer (17–19). Additionally, the overexpression of cyclin E correlates with worse outcome in patients with breast cancer, rectal cancer and gastrointestinal cancer (20–22). Some evidence has shown that low expression of cyclin F may be tumorigenic. It has been proposed that the downregulation of cyclin F promotes centrosomal and mitotic abnormalities associated with impaired degradation of CP110, an important centriolar protein (23). Moreover, cyclin F-mediated degradation of CDC-6 suppresses genome instability and prevents re-replication, limiting the number of cells with DNA content greater than 4N (24). Pan et al showed that different levels of cyclin F, cyclin D and RBL1 between 2D and 3D cultured cells may be associated with radioresistance of cells in 3-dimensional culture. They noted that A549 cells cultured in 3D exhibited lower levels of cyclin F and were less susceptible to G2/M cell cycle arrest after X-ray irradiation (25). However, the potential role of cyclin F as a tumor-promoting factor and the underlying mechanism remain elusive. The Oct4/NIPP1-CCNF/PP1 axis is responsible for maintenance of retinoblastoma protein 1 (Rb1) in the hyperphosphorylated state providing stem cell self-renewal and increased proliferation. Inactivation of Rb1 via CCNF/PP1 is also associated with enhanced ovarian cancer aggressiveness (26,27). In our pathway analysis, we observed a decrease in the cell signaling-related pathway activity and increase in the cell cycle-related pathways in patients with upregulated levels of cyclin F. A recent report showed that cyclin F is a bridge between AKT kinase and cell cycle machinery. Choudhury et al hypothesized a model where growth signaling initiates a positive loop where AKT phosphorylates and stabilizes cyclin F in the SCF complex. This stabilization inhibits degradation of cyclin F via APC/C (Cdh1) complex and promotes SCF-dependent degradation of Cdh1. Degradation of Cdh1 is essential for S phase entry and loss of cyclin F impairs cell cycle progression (28,29). Activation of the PI3K/AKT pathway is a common event in a variety of cancer diseases and it is believed to contribute to drug resistance. Although, we did not observe clear symptoms of PI3K/AKT activation, our analysis revealed downregulated INPP4B, tumor suppressor antagonizing PI3K/AKT pathway. Loss of INPP4B was found to increase AKT activation and drive higher proliferation rate and metastasis (30). It has been also reported that a decreased level of INPP4B is releted to higher proliferative, invasive and metastatic potential of melanocytic neoplasms (31). In contradiction to these reports is a study by Chi et al where upregulation of INPP4B in a melanoma subset was observed. Furthermore, INPP4B driven proliferation was Akt-independent and was mediated by serum- and glucocorticoid-regulated kinase 3 (SGK3). Additionally, they observed no significant differences between primary and metastatic melanoma suggesting the involvement of INPP4B in developing cancer from the early stages (32).
In the present study, the upregulation of cyclin F mRNA was found to coincide with the downregulation of p27 protein, important cell cycle regulator involved in G1 arrest. Akman et al found that patients with melanoma are characterized by lower p27 expression in comparison to patients with benign nevi and dysplastic nevi (33). Furthermore, Florenes et al reported that decreased expression of p27 is associated with increasing Breslow thickness and lower disease-free survival rates in primary nodular melanoma (34). Additionally, the low expression of p27 in melanocytic lesions may be responsible for its high proliferation rate (35). The lack of proper control in regards to cell cycle events is typical for cancer cells. As was mentioned in the introduction, the overexpression of cyclins is very common in various malignancies. In our analysis, elevated levels of cyclin F mRNA were also associated with upregulation of cyclin E1 and B1 proteins. Elevated levels of cyclin E1 were observed in melanoma and enhanced expression of cyclin E was noted in both primary and metastatic melanomas. In contrast, its overexpression was not observed in non-malignant nevi (36). Bales et al reported that cyclin E is overexpressed in melanoma and present in the low-molecular form. Noteworthy, transfection of a primary cutaneous melanoma cell line with low tumorigenic and metastatic potential with low-molecular cyclin E forms resulted in the development of angiogenic tumors with prominent perineural invasion. Additionally, truncated forms of cyclin E triggered a dramatic increase in a number of metastasis events (37). In turn, cyclin B1 is involved in proliferation and metastatic potential of melanoma cells (38). Silencing of cyclin B exerts an antitumor effect on melanoma cells and lung metastases, both in vitro and in vivo (39).
Kruiswijk et al reported that patients with elevated levels of cyclin B1, Pin1 and FOXM1 display a worse outcome and exhibit increased mortality (40). FOXM1 is a pro-proliferative and pro-survival transcription factor participating in DNA repair. Moreover, these data are in agreement with our analysis, where a significant increase in FOXM1 protein in patients with upregulated cyclin F mRNA was noted. It suggests possible activation of cyclin F expression by FOXM1, but further research is needed to clarify this. Moreover, the upregulation of FOXM1 coincides with downregulation of FOXO3a. The abrogation of FOXO3a function was found to lead to increased tumor aggressiveness in melanoma and renal carcinoma (41,42). Another important observation made in this study is that 4E-BP1 (4E binding protein 1) was hyperphosphorylated in patients with upregulated cyclin expression. Phosphorylation of 4E-BP1 results in dissociation from translation factor eIF4E and allows cap-dependent translation. Phospho-4E-BP1 may also be useful as a marker of mTOR pathway activity and integrates signals obtained from PI3K/AKT and RAS/RAF/MEK/ERK pathways (43). Additionally, concomitant hyperphosphorylation of 4E-BP1 and activation of the PI3K/AKT pathway results in resistance to mTOR inhibitors. Moreover, in hypoxic conditions, 4E-BP1 initiates translation of proteins responsible for angiogenesis (VEGF-A), hypoxia response (HIF1α) and apoptosis resistance (Bcl-2) in advanced cancer (44,45). Increased levels of phosphorylated 4E-BP1 are also associated with poor overall survival and significant difference in post-recurrence survival (46). It is possible that cyclin F is a part of the specific cellular environment, promoting cell proliferation and survival.
The ability of cancer cells to efficiently repair DNA is a significant barrier to successful treatment. RRM2 is a part of the RNR and has been reported to be partially responsible for chemoresistance of cancer cells, including melanoma. However, our analysis did not reveal significant changes in overall survival or disease-free survival between patients with differential RRM2 mRNA expression. Aird et al showed that high RRM2 expression is correlated with worse outcome in melanoma patients (8). Silencing of RRM2 inhibited melanoma growth which suggests the involvement of RRM2 in melanoma progression. Silencing of RRM2 and treatment with mutant BRAF inhibitor PLX4720 simultaneously and synergistically inhibited melanoma growth (11). It is possible that the negative effect of RRM2 overexpression is limited to patients bearing BRAFV600E mutation, but we cannot confirm this using TCGA data due to an insufficient number of patients with the BRAF mutation in the cohort.
Beyond controlling RRM2 levels, cyclin F is a limiting factor in histone H2.AX signalization. In the G2 phase cyclin F mediates degradation of SLBP protein which promotes synthesis of H2AFX mRNA. Presence of SLBP in the G2 phase increases H2.AX levels and makes the cell more susceptible to apoptosis under genotoxic stress. It is another piece of evidence showing how cyclin F promotes cancer progression (47). Moreover, we observed an alteration in expression of other DNA-repair related proteins: XRCC1, PARP1, PCNA, and MSH6. All proteins were upregulated which is a hallmark of efficient DNA repair systems and a potential obstacle to successful treatment. However, the prognostic status of XRCC1 is ambiguous. Its overexpression is associated with less favorable prognosis in head and neck squamous carcinoma. Decreased levels of XRCC1 are responsible for acute side-effects after radiotherapy in breast cancer patients. Loss of XRCC1 confers a more aggressive phenotype in melanoma (48–50). It suggests an indirect effect of cyclin F overexpression on the DNA damage repair system. Additionally, PCNA in patients with upregulated cyclin F is very significantly increased, what confirms the higher proliferation potential of cells overexpressing cyclin F. These findings confirm a study by Wang et al in which treatment of cells with stimulatory polysaccharides from abalone, significantly increased the expression of cyclin B1, CDK1 and cyclin F (51).
Another interesting observation was increased expression of TFRC (transferrin receptor 1) gene in patients with high expression of cyclin F and RRM2 mRNA. It has been reported that melanoma cells are able to upregulate transferrin receptor 1 through the hyaluronan/CD44 pathway. It is possible that this pathway promotes proliferation providing alternative iron supply for melanoma cells. High expression of TFRC is associated with unfavorable prognosis in breast and pancreatic cancer (52–54).
This newly discovered relationship between mRNA expression of CCNF and RRM2 provide and attractive point for further investigations in the field of dermato-oncology. Our analysis was performed using independent data obtained from TCGA and provide many key results that can be used in further explanation of the precise mechanisms. Moreover, we expect that the present results will be useful to other researchers and induce further investigations, essential for better diagnosis, prediction, therapy response, but also for better selection of patients for optimal therapy against skin melanoma. A high number of clones contributes to an exceptional level of intratumor heterogeneity of melanoma, but also refers to metastases which may originate from different subclones of the primary tumor. This creates an obstacle to proper diagnosis and successful treatment (55). Increased research on the topic is needed for understanding the limitation or failure of contemporary therapies and the precise mechanism must and will be elucidated by our team in vitro in the immediate future using melanoma cancer cell panels. We suggest here to investigate the precise mechanism indicated in the study using all following cell lines: SK-MEL-1, A375, G-361, SK-MEL-3, SH-4, SK-MEL-24, RPMI-7951. However, we hope that the publication of in silico analyses accelerates the development and inspires other scientific teams to conduct similar research in the field.
In conclusion, the present study is a first attempt to elucidate the influence of cyclin F mRNA expression on the outcome of melanoma patients. High expression of cyclin F mRNA is associated with worse overall survival. Moreover, in silico analysis revealed that upregulated cyclin F mRNA expression is associated with activation of molecular pathways responsible for melanoma proliferation, metastatic potential and survival. These findings are a good starting point to address new cyclin F targets and interactions which drive the increased aggressiveness of the tumor.
Acknowledgements
Not applicable.
Funding
This study was supported by a grant from the National Science Centre, Poland (grant no. 2016/21/B/NZ7/01121 to AG).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
MG and AG designed the study. MG and AK performed the analyses, interpreted the data and wrote the study. DG and AG revised manuscript critically for important intellectual content. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The present study was approved by the Bioethics Committee of the Nicolaus Copernicus University in Toruń functioning at Collegium Medicum in Bydgoszcz (KB 554/2016).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Niezgoda A, Niezgoda P, Czajkowski R. Novel approaches to treatment of advanced melanoma: A review on targeted therapy and immunotherapy. BioMed Res Int. 2015;2015:851387. doi: 10.1155/2015/851387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson DB, Sosman JA. Therapeutic advances and treatment options in metastatic melanoma. JAMA Oncol. 2015;1:380–386. doi: 10.1001/jamaoncol.2015.0565. [DOI] [PubMed] [Google Scholar]
- 3.Maverakis E, Cornelius LA, Bowen GM, Phan T, Patel FB, Fitzmaurice S, He Y, Burrall B, Duong C, Kloxin AM, et al. Metastatic melanoma - a review of current and future treatment options. Acta Derm Venereol. 2015;95:516–524. doi: 10.2340/00015555-2035. [DOI] [PubMed] [Google Scholar]
- 4.Galper J, Rayner SL, Hogan AL, Fifita JA, Lee A, Chung RS, Blair IP, Yang S. Cyclin F: A component of an E3 ubiquitin ligase complex with roles in neurodegeneration and cancer. Int J Biochem Cell Biol. 2017;89:216–220. doi: 10.1016/j.biocel.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 5.D'Angiolella V, Donato V, Forrester FM, Jeong YT, Pellacani C, Kudo Y, Saraf A, Florens L, Washburn MP, Pagano M. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 2012;149:1023–1034. doi: 10.1016/j.cell.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grolmusz VK, Karászi K, Micsik T, Tóth EA, Mészáros K, Karvaly G, Barna G, Szabó PM, Baghy K, Matkó J, et al. Cell cycle dependent RRM2 may serve as proliferation marker and pharmaceutical target in adrenocortical cancer. Am J Cancer Res. 2016;6:2041–2053. [PMC free article] [PubMed] [Google Scholar]
- 7.Han P, Lin Z-R, Xu L-H, Zhong Q, Zhu XF, Liang FY, Cai Q, Huang XM, Zeng MS. Ribonucleotide reductase M2 subunit expression and prognostic value in nasopharyngeal carcinoma. Mol Med Rep. 2015;12:401–409. doi: 10.3892/mmr.2015.3360. [DOI] [PubMed] [Google Scholar]
- 8.Aird KM, Zhang G, Li H, Tu Z, Bitler BG, Garipov A, Wu H, Wei Z, Wagner SN, Herlyn M, et al. Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell Reports. 2013;3:1252–1265. doi: 10.1016/j.celrep.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mah V, Alavi M, Márquez-Garbán DC, Maresh EL, Kim SR, Horvath S, Bagryanova L, Huerta-Yepez S, Chia D, Pietras R, et al. Ribonucleotide reductase subunit M2 predicts survival in subgroups of patients with non-small cell lung carcinoma: Effects of gender and smoking status. PLoS One. 2015;10:e0127600. doi: 10.1371/journal.pone.0127600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fatkhutdinov N, Sproesser K, Krepler C, Liu Q, Brafford PA, Herlyn M, Aird KM, Zhang R. Targeting RRM2 and mutant BRAF is a novel combinatorial strategy for melanoma. Mol Cancer Res. 2016;14:767–775. doi: 10.1158/1541-7786.MCR-16-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuckerman JE, Hsueh T, Koya RC, Davis ME, Ribas A. siRNA knockdown of ribonucleotide reductase inhibits melanoma cell line proliferation alone or synergistically with temozolomide. J Invest Dermatol. 2011;131:453–460. doi: 10.1038/jid.2010.310. [DOI] [PubMed] [Google Scholar]
- 13.Fu J, Qiu H, Cai M, Pan Y, Cao Y, Liu L, Yun J, Zhang CZ. Low cyclin F expression in hepatocellular carcinoma associates with poor differentiation and unfavorable prognosis. Cancer Sci. 2013;104:508–515. doi: 10.1111/cas.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun X, Zhangyuan G, Shi L, Wang Y, Sun B, Ding Q. Prognostic and clinicopathological significance of cyclin B expression in patients with breast cancer: A meta-analysis. Medicine (Baltimore) 2017;96:e6860. doi: 10.1097/MD.0000000000006860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W, Dong Q, Li L, Zhang Z, Cai X, Pan X. Prognostic significance of claudin-1 and cyclin B1 protein expression in patients with hypopharyngeal squamous cell carcinoma. Oncol Lett. 2016;11:2995–3002. doi: 10.3892/ol.2016.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang Y, Liang X, Jiang W, Li J, Xu J, Cai X. Cyclin b1 suppresses colorectal cancer invasion and metastasis by regulating e-cadherin. PLoS One. 2015;10:e0126875. doi: 10.1371/journal.pone.0126875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shan YS, Hsu HP, Lai MD, Hung YH, Wang CY, Yen MC, Chen YL. Cyclin D1 overexpression correlates with poor tumor differentiation and prognosis in gastric cancer. Oncol Lett. 2017;14:4517–4526. doi: 10.3892/ol.2017.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin RJ, Lubpairee T, Liu KY, Anderson DW, Durham S, Poh CF. Cyclin D1 overexpression is associated with poor prognosis in oropharyngeal cancer. J Otolaryngol Head Neck Surg. 2013;42:23. doi: 10.1186/1916-0216-42-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahlin C, Lundgren C, Embretsén-Varro E, Jirström K, Blomqvist C, Fjällskog ML. High expression of cyclin D1 is associated to high proliferation rate and increased risk of mortality in women with ER-positive but not in ER-negative breast cancers. Breast Cancer Res Treat. 2017;164:667–678. doi: 10.1007/s10549-017-4294-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luhtala S, Staff S, Tanner M, Isola J. Cyclin E amplification, over-expression, and relapse-free survival in HER-2-positive primary breast cancer. Tumour Biol. 2016;37:9813–9823. doi: 10.1007/s13277-016-4870-z. [DOI] [PubMed] [Google Scholar]
- 21.Zhou YJ, Xie YT, Gu J, Yan L, Guan GX, Liu X. Overexpression of cyclin E isoforms correlates with poor prognosis in rectal cancer. Eur J Surg Oncol. 2011;37:1078–1084. doi: 10.1016/j.ejso.2011.08.139. [DOI] [PubMed] [Google Scholar]
- 22.Huang L, Ren F, Tang R, Feng Z, Chen G. Prognostic value of expression of cyclin E in gastrointestinal cancer: A systematic review and meta-analysis. Technol Cancer Res Treat. 2016;15:12–19. doi: 10.1177/1533034614568098. [DOI] [PubMed] [Google Scholar]
- 23.D'Angiolella V, Donato V, Vijayakumar S, Saraf A, Florens L, Washburn MP, Dynlacht B, Pagano M. SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138–142. doi: 10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walter D, Hoffmann S, Komseli E-S, Rappsilber J, Gorgoulis V, Sørensen CS. SCF(Cyclin F)-dependent degradation of CDC6 suppresses DNA re-replication. Nat Commun. 2016;7:10530. doi: 10.1038/ncomms10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan D, Chen Y, Du Y, Ren Z, Li X, Hu B. Methylation of promoter of RBL1 enhances the radioresistance of three dimensional cultured carcinoma cells. Oncotarget. 2017;8:4422–4435. doi: 10.18632/oncotarget.12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schoeftner S, Scarola M, Comisso E, Schneider C, Benetti R. An Oct4-pRb axis, controlled by MiR-335, integrates stem cell self-renewal and cell cycle control. Stem Cells. 2013;31:717–728. doi: 10.1002/stem.1315. [DOI] [PubMed] [Google Scholar]
- 27.Comisso E, Scarola M, Rosso M, Piazza S, Marzinotto S, Ciani Y, Orsaria M, Mariuzzi L, Schneider C, Schoeftner S, et al. OCT4 controls mitotic stability and inactivates the RB tumor suppressor pathway to enhance ovarian cancer aggressiveness. Oncogene. 2017;36:4253–4266. doi: 10.1038/onc.2017.20. [DOI] [PubMed] [Google Scholar]
- 28.Choudhury R, Bonacci T, Wang X, Truong A, Arceci A, Zhang Y, Mills CA, Kernan JL, Liu P, Emanuele MJ. The E3 ubiquitin ligase SCF(cyclin F) transmits AKT signaling to the cell-cycle machinery. Cell Reports. 2017;20:3212–3222. doi: 10.1016/j.celrep.2017.08.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choudhury R, Bonacci T, Arceci A, Lahiri D, Mills CA, Kernan JL, Branigan TB, DeCaprio JA, Burke DJ, Emanuele MJ. APC/C and SCF(cyclin F) constitute a reciprocal feedback circuit controlling S-phase entry. Cell Reports. 2016;16:3359–3372. doi: 10.1016/j.celrep.2016.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Chew C, Lunardi A, Gulluni F, Ruan DT, Chen M, Salmena L, Nishino M, Papa A, Ng C, Fung J, et al. In vivo role of INPP4B in tumor and metastasis suppression through regulation of PI3K-AKT signaling at endosomes. Cancer Discov. 2015;5:740–751. doi: 10.1158/2159-8290.CD-14-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez-Lorenzo R, Gill KZ, Shen C-H, Zhao FX, Zheng B, Schulze HJ, Silvers DN, Brunner G, Horst BA. A tumor suppressor function for the lipid phosphatase INPP4B in melanocytic neoplasms. J Invest Dermatol. 2014;134:1359–1368. doi: 10.1038/jid.2013.511. [DOI] [PubMed] [Google Scholar]
- 32.Chi MN, Guo ST, Wilmott JS, Guo XY, Yan XG, Wang CY, Liu XY, Jin L, Tseng HY, Liu T, et al. INPP4B is upregulated and functions as an oncogenic driver through SGK3 in a subset of melanomas. Oncotarget. 2015;6:39891–39907. doi: 10.18632/oncotarget.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akman A, Ciftcioglu MA, Ozbey C, Alpsoy E. Expression of cell cycle inhibitor p27Kip1 in nevi and melanomas. Indian J Dermatol Venereol Leprol. 2008;74:551. doi: 10.4103/0378-6323.44341. [DOI] [PubMed] [Google Scholar]
- 34.Flørenes VA, Maelandsmo GM, Kerbel RS, Slingerland JM, Nesland JM, Holm R. Protein expression of the cell-cycle inhibitor p27Kip1 in malignant melanoma: Inverse correlation with disease-free survival. Am J Pathol. 1998;153:305–312. doi: 10.1016/S0002-9440(10)65572-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivan D, Diwan AH, Esteva FJ, Prieto VG. Expression of cell cycle inhibitor p27Kip1 and its inactivator Jab1 in melanocytic lesions. Mod Pathol. 2004;17:811–818. doi: 10.1038/modpathol.3800123. [DOI] [PubMed] [Google Scholar]
- 36.Georgieva J, Sinha P, Schadendorf D. Expression of cyclins and cyclin dependent kinases in human benign and malignant melanocytic lesions. J Clin Pathol. 2001;54:229–235. doi: 10.1136/jcp.54.3.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bales E, Mills L, Milam N, McGahren-Murray M, Bandyopadhyay D, Chen D, Reed JA, Timchenko N, van den Oord JJ, Bar-Eli M, et al. The low molecular weight cyclin E isoforms augment angiogenesis and metastasis of human melanoma cells in vivo. Cancer Res. 2005;65:692–697. [PubMed] [Google Scholar]
- 38.Lu M, Breyssens H, Salter V, Zhong S, Hu Y, Baer C, Ratnayaka I, Sullivan A, Brown NR, Endicott J, et al. Restoring p53 function in human melanoma cells by inhibiting MDM2 and cyclin B1/CDK1-phosphorylated nuclear iASPP. Cancer Cell. 2013;23:618–633. doi: 10.1016/j.ccr.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 39.Kedinger V, Meulle A, Zounib O, Bonnet ME, Gossart JB, Benoit E, Messmer M, Shankaranarayanan P, Behr JP, Erbacher P, et al. Sticky siRNAs targeting survivin and cyclin B1 exert an antitumoral effect on melanoma subcutaneous xenografts and lung metastases. BMC Cancer. 2013;13:338. doi: 10.1186/1471-2407-13-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kruiswijk F, Hasenfuss SC, Sivapatham R, Baar MP, Putavet D, Naipal KA, van den Broek NJ, Kruit W, van der Spek PJ, van Gent DC, et al. Targeted inhibition of metastatic melanoma through interference with Pin1-FOXM1 signaling. Oncogene. 2016;35:2166–2177. doi: 10.1038/onc.2015.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ni D, Ma X, Li H-Z, Gao Y, Li XT, Zhang Y, Ai Q, Zhang P, Song EL, Huang QB, et al. Downregulation of FOXO3a promotes tumor metastasis and is associated with metastasis-free survival of patients with clear cell renal cell carcinoma. Clin Cancer Res. 2014;20:1779–1790. doi: 10.1158/1078-0432.CCR-13-1687. [DOI] [PubMed] [Google Scholar]
- 42.Zanella F, Renner O, García B, Callejas S, Dopazo A, Peregrina S, Carnero A, Link W. Human TRIB2 is a repressor of FOXO that contributes to the malignant phenotype of melanoma cells. Oncogene. 2010;29:2973–2982. doi: 10.1038/onc.2010.58. [DOI] [PubMed] [Google Scholar]
- 43.Ayuso MI, Hernández-Jiménez M, Martín ME, Salinas M, Alcázar A. New hierarchical phosphorylation pathway of the translational repressor eIF4E-binding protein 1 (4E-BP1) in ischemia-reperfusion stress. J Biol Chem. 2010;285:34355–34363. doi: 10.1074/jbc.M110.135103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sherrill KW, Byrd MP, Van Eden ME, Lloyd RE. BCL-2 translation is mediated via internal ribosome entry during cell stress. J Biol Chem. 2004;279:29066–29074. doi: 10.1074/jbc.M402727200. [DOI] [PubMed] [Google Scholar]
- 45.Qin X, Jiang B, Zhang Y. 4E-BP1, a multifactor regulated multifunctional protein. Cell Cycle. 2016;15:781–786. doi: 10.1080/15384101.2016.1151581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Reilly KE, Warycha M, Davies MA, Rodrik V, Zhou XK, Yee H, Polsky D, Pavlick AC, Rosen N, Bhardwaj N, et al. Phosphorylated 4E-BP1 is associated with poor survival in melanoma. Clin Cancer Res. 2009;15:2872–2878. doi: 10.1158/1078-0432.CCR-08-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dankert JF, Rona G, Clijsters L, Geter P, Skaar JR, Bermudez-Hernandez K, Sassani E, Fenyö D, Ueberheide B, Schneider R, et al. Cyclin F-mediated degradation of SLBP limits H2A.X accumulation and apoptosis upon genotoxic stress in G2. Mol Cell. 2016;64:507–519. doi: 10.1016/j.molcel.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ang M-K, Patel MR, Yin X-Y, Sundaram S, Fritchie K, Zhao N, Liu Y, Freemerman AJ, Wilkerson MD, Walter V, et al. High XRCC1 protein expression is associated with poorer survival in patients with head and neck squamous cell carcinoma. Clin Cancer Res. 2011;17:6542–6552. doi: 10.1158/1078-0432.CCR-10-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Batar B, Guven G, Eroz S, Bese NS, Guven M. Decreased DNA repair gene XRCC1 expression is associated with radiotherapy-induced acute side effects in breast cancer patients. Gene. 2016;582:33–37. doi: 10.1016/j.gene.2016.01.040. [DOI] [PubMed] [Google Scholar]
- 50.Bhandaru M, Martinka M, Li G, Rotte A. Loss of XRCC1 confers a metastatic phenotype to melanoma cells and is associated with poor survival in patients with melanoma. Pigment Cell Melanoma Res. 2014;27:366–375. doi: 10.1111/pcmr.12212. [DOI] [PubMed] [Google Scholar]
- 51.Wang YM, Wu FJ, Du L, Li GY, Takahashi K, Xue Y, Xue CH. Effects of polysaccharides from abalone (Haliotis discus hannai Ino) on HepG2 cell proliferation. Int J Biol Macromol. 2014;66:354–361. doi: 10.1016/j.ijbiomac.2014.01.032. [DOI] [PubMed] [Google Scholar]
- 52.Miller LD, Coffman LG, Chou JW, Black MA, Bergh J, D'Agostino R, Jr, Torti SV, Torti FM. An iron regulatory gene signature predicts outcome in breast cancer. Cancer Res. 2011;71:6728–6737. doi: 10.1158/0008-5472.CAN-11-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laube F, Glanz D. Modulation of Melanotransferrin and Transferrin Receptor 1 (TFRC)- and CD44-based Signaling for TFRC Up-regulation in Human Melanoma Cells. Anticancer Res. 2017;37:3001–3007. doi: 10.21873/anticanres.11654. [DOI] [PubMed] [Google Scholar]
- 54.Ryschich E, Huszty G, Knaebel HP, Hartel M, Büchler MW, Schmidt J. Transferrin receptor is a marker of malignant phenotype in human pancreatic cancer and in neuroendocrine carcinoma of the pancreas. Eur J Cancer. 2004;40:1418–1422. doi: 10.1016/j.ejca.2004.01.036. [DOI] [PubMed] [Google Scholar]
- 55.Grzywa TM, Paskal W, Włodarski PK. Intratumor and Intertumor Heterogeneity in Melanoma. Transl Oncol. 2017;10:956–975. doi: 10.1016/j.tranon.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.