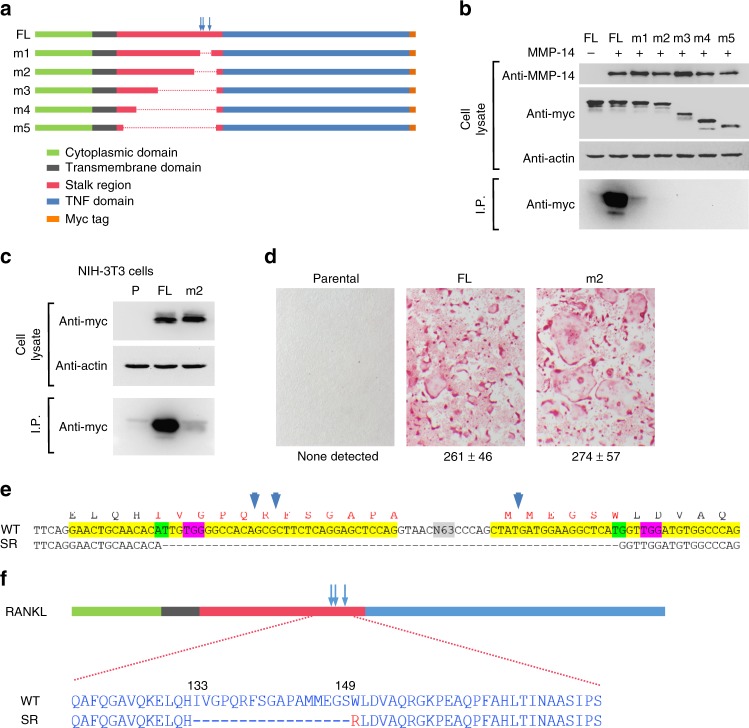
Fig. 1.
Development of a sheddase-resistant RANKL. a Diagram of full-length (FL) and mutant RANKL constructs. Known cleavage sites are indicated by vertical arrows. b Sheddase assay in transiently transfected 293T cells. This experiment was replicated at least once. c Sheddase assay in stably transduced NIH-3T3 cells. P parental NIH-3T3 cells. This experiment was replicated at least once. d Osteoclast formation in co-cultures of bone marrow macrophages with the indicated NIH-3T3 cells shown in “c”. Osteoclasts are stained red. Quantification of multinucleated osteoclasts from triplicate wells shown below, mean ± s.d. This experiment was replicated at least once. e Partial DNA sequence of murine Tnfsf11 gene showing CRISPR/Cas9 gene-editing strategy. Exon 3 and a portion of exon 4 are highlighted in yellow. The sequence of wild-type (WT) and sheddase-resistant (SR) mutants are shown. N63 indicates 63 bp of intron 3 not shown. The PAM sequences for each of the two single-guide RNA (sgRNA) targets are highlighted in pink and their respective cut sites highlighted in green. Amino acids encoded by the exons are shown above the DNA sequence with the locations of known proteolytic cleavage sites indicated by the vertical blue arrows. The SR mutant was created by non-homologous end-joining of the double-stranded DNA cuts directed by the sgRNAs. f Diagram of a portion of the WT and SR RANKL proteins highlighting the amino acid sequences in the cleavage region. The red R indicates the position of an arginine resulting from deletion of the sequence shown in “e”