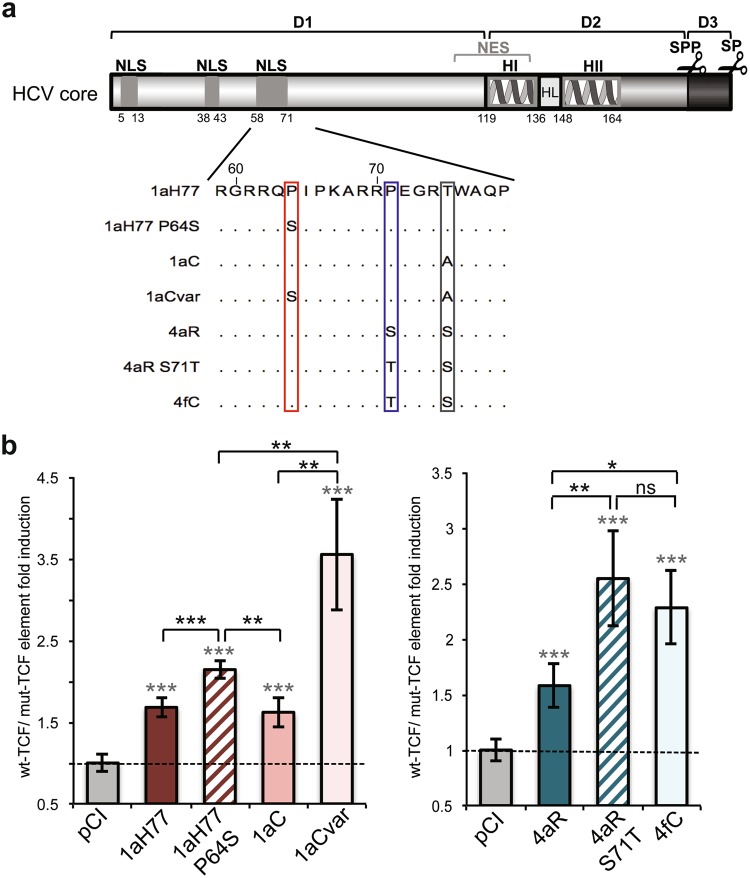
Figure 2.
Involvement of residues at position 64 and 71 of HCV core in the modulation of TCF-element activation. (a) HCV core is schematically represented with its subdomains (D1, D2, D3) including predicted or experimentally-demonstrated nuclear localization signals (NLS), nuclear export signals (NES), alpha-helices (H), loop between alpha-helices (HL), and location of cleavage by cellular signal peptidase (SP) and signal peptide peptidase (SPP) (scissors). Numbering below the scheme corresponds to amino acid position framing each element in core. An alignment of core sequences from clinical isolates and engineered mutant derivatives 1aH77 P64S and 4aR S71T is shown in the blown-up below. Amino acid differences are boxed. (b,c) Relative TCF element activation in HEK293 cells expressing natural or engineered core variants and statistical analyses were established and represented as described in Fig. 1 caption (means ± SD of quintuplicates obtained in 3 independent experiments).