FIG 1 .
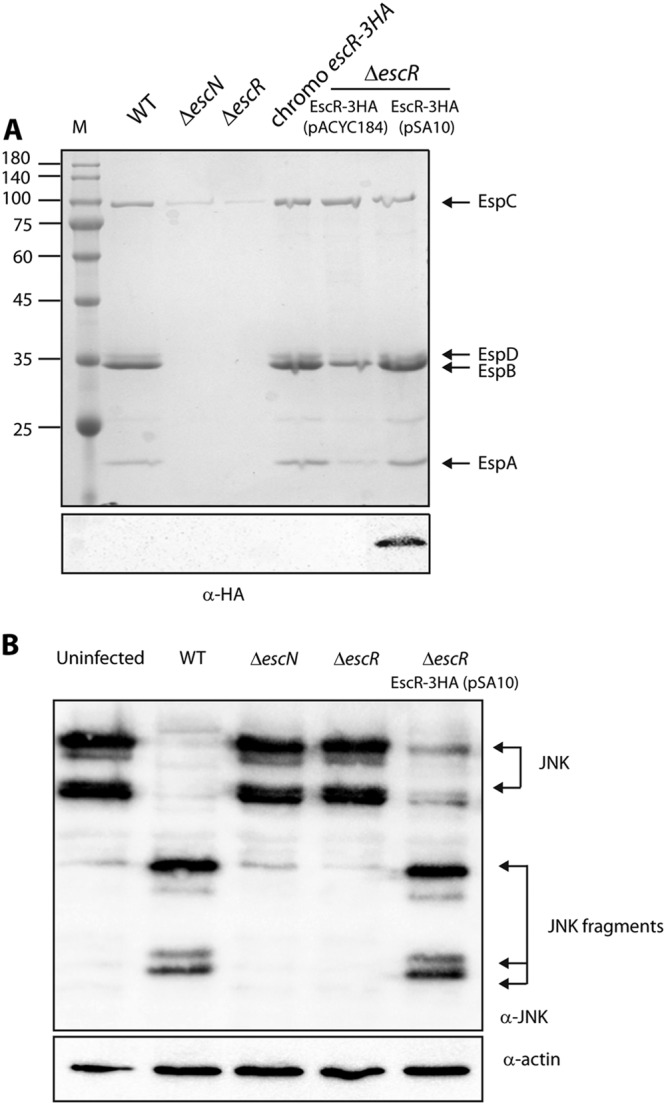
C-terminally labeled EscR can complement the ΔescR mutant. (A) Protein secretion profiles of EPEC strains grown under T3SS-inducing conditions: wild type (WT), ΔescN mutant (a T3SS ATPase mutant), ΔescR mutant, chromosomal escR-3HA strain, and ΔescR mutant complemented with EscR-3HA (encoded on either pACYC184 or pSA10). The secreted proteins were concentrated from supernatants of bacterial cultures and analyzed by SDS-PAGE and Coomassie blue staining. The T3SS-secreted translocators EspA, EspB, and EspD are indicated on the right of the gel. Also indicated is the location of EspC, which is not secreted by the T3SS. For the ΔescN and ΔescR strains, no T3SS activity was observed. Strains carrying the chromosomal escR-3HA or the plasmids encoding EscR-3HA showed proper T3SS activity. The expression of EscR-3HA was examined by analyzing the bacterial pellets by SDS-PAGE and Western blot analysis with an anti-HA antibody. EscR-3HA expression was detected only for EscR-3HA expressed from a high-copy-number plasmid, pSA10. Numbers at left are molecular masses in kilodaltons. (B) To examine whether pEscR-3HA can complement the ΔescR mutant in a host cell infection model, HeLa cells were infected with EPEC strains: WT, ΔescN, and ΔescR strains and ΔescR strain complemented with pEscR-3HA. After 3 h, cells were washed, and host cell proteins were extracted and subjected to Western blot analysis using anti-JNK and anti-actin (loading control) antibodies. JNK and its degradation fragments are indicated at the right of the gel. WT EPEC showed massive degradation of JNK compared to the uninfected sample and the samples infected with ΔescN or ΔescR mutant strains. The ΔescR strain complemented with pEscR-3HA showed a similar JNK degradation profile as WT EPEC, thus suggesting that pEscR-3HA is functional and therefore fully complements the ΔescR infection phenotype.
