FIG 1.
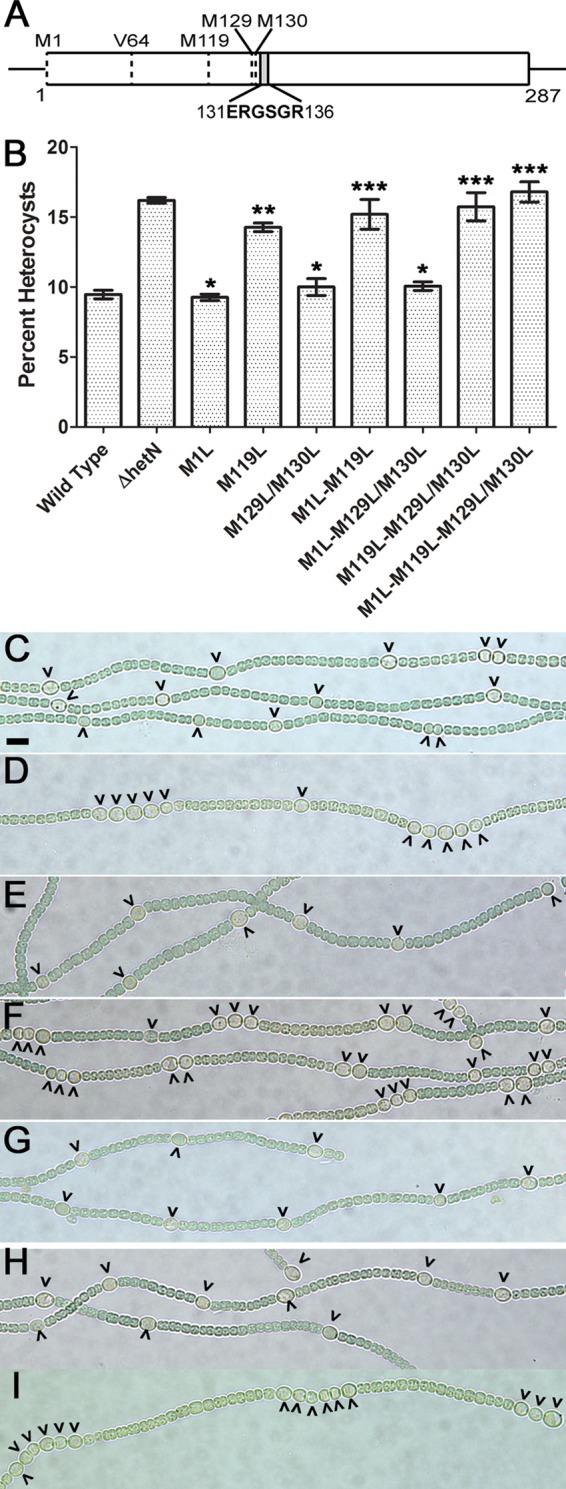
Alleles of hetN encoding M119L substitutions result in an Mch phenotype similar to that of a ΔhetN strain. (A) Schematic depicting the positions of potential start codons (M1, M119, M129/130, and V64, encoded by a GTG codon) and the ERGSGR motif in the hetN coding region. (B) Heterocyst percentages for the wild-type and ΔhetN strains as well as strains with the indicated chromosomal mutations. Data are presented as the averages of results from three replicates. Error bars represent standard deviations. Statistical significance was calculated by a t test with a P value of <0.05 (*, different from the ΔhetN strain and not different from the wild type; **, different from both the ΔhetN and wild-type strains; ***, different from the wild type and not different from the ΔhetN strain). (C to I) Bright-field images of the wild type (C), UHM150 (ΔhetN) (D), UHM328 [hetN(M1L)] (E), UHM345 [hetN(M119L)] (F), UHM346 [hetN(M129L/M130L)] (G), UHM347 [hetN(M1L,M129L/M130L)] (H), and UHM349 [hetN(M1L,M119L,M129L/M130L)] (I). Micrographs were taken 48 h after the removal of combined nitrogen. Carets indicate heterocysts. Bar in panel C, 10 μm.
