Fig. 3.
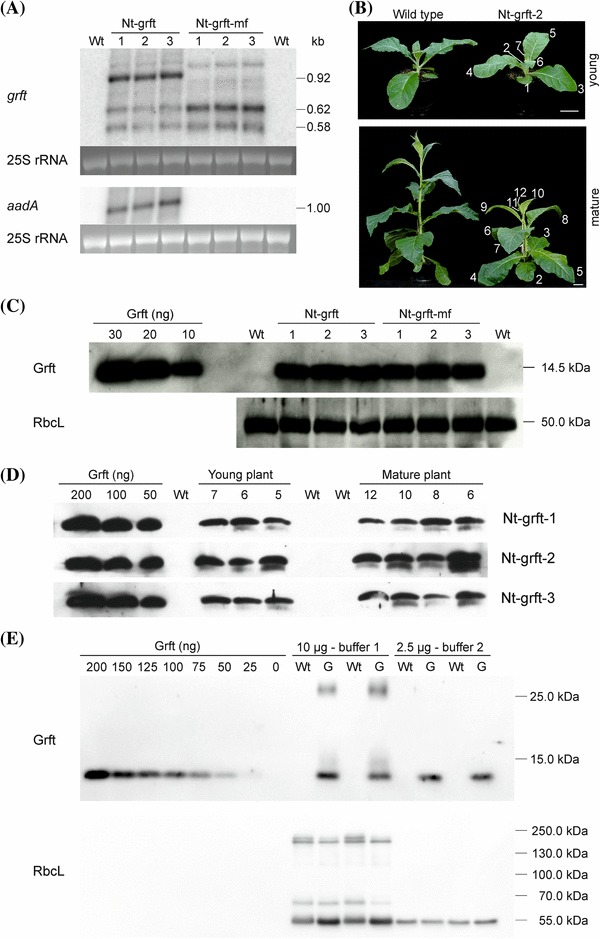
Expression of griffithsin in transplastomic tobacco plants. a Accumulation of grft and aadA transcripts. The grft-specific probe detects three major transcript species in Nt-grft plants (~ 0.92, ~ 0.62, ~ 0.58 kb) and two major transcripts in Nt-grft-mf plants (~ 0.62, ~ 0.58 kb), a pattern observed for the pKP9 expression cassette in previous studies (e.g., Zhou et al. 2008). The ~ 1 kb aadA transcript accumulates in Nt-grft plants, but is not detectable in the wild type (Wt) and in marker gene-free plants (Nt-grft-mf). The ethidium bromide-stained 25S rRNA of the cytosolic 80S ribosomes served as a loading control. b A transplastomic plant (Nt-grft-2) and a wild type grown under standard greenhouse conditions for six (young) and eight (mature) weeks to illustrate the sampling of leaves (consecutively numbered from the bottom to the top of the plant) for immunoblot analyses. Scale bar: 5 cm. c Accumulation of the griffithsin (Grft) protein in seedlings. The griffithsin-specific antibody detects a protein of the expected size (14.5 kDa) in total soluble protein extracts (5 µg loaded per lane) of all transplastomic plants. Immunodetection of RbcL served as loading control. d Comparison of griffithsin accumulation in a developmental series of leaves. Immunodetection of griffithsin in total soluble protein extracts (5 µg per sample) of three different leaves from a young plant (leaves number 7, 6 and 5) and four leaves from a mature plant (leaves 12, 10, 8 and 6). e Determination of griffithsin accumulation levels in total soluble protein (TSP) samples. TSP was extracted with two different buffers (see “Materials and methods”) from the wild type (Wt) and the Nt-grft-mf-2 line (G). Two replicates per buffer and plant line were analyses. For buffer 1, samples of 10 µg TSP and for buffer 2, samples of 2.5 µg TSP were loaded. Immunodetection of RbcL served as loading control. In buffer 1, the griffithsin dimer is detected and RbcL shows larger complexes, suggesting incomplete protein denaturation. Using known standards of recombinant histidine-tagged griffithsin, griffithsin was detected at approximately 2% of TSP with buffer 1 (including the dimer) and approximately 5% with buffer 2
