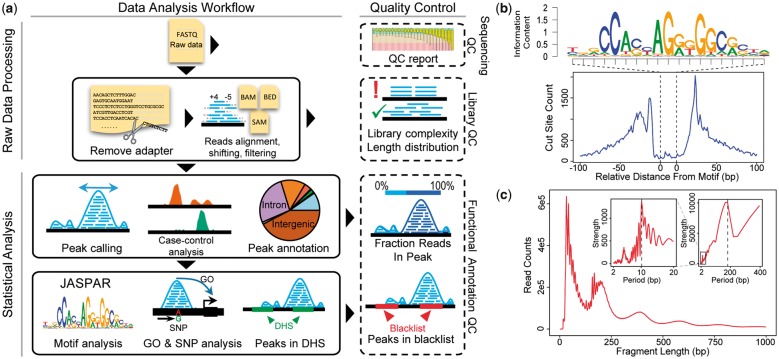
Fig. 1.
(a) esATAC workflow. esATAC pipeline is mainly divided into two parts, raw data processing and statistical analysis. QC functions at multiple levels are provided, including sequencing QC, library QC and functional annotation QC. (b) and (c) Examples of analyzing ATAC-seq data (GEO accession number GSE47753, see Supplementary Material). (b) CTCF footprinting. (c) Fragment length distribution. Periodicity of approximately 200 base pairs (bp) for nucleosome protection and 10.4 bp for the pitch of the DNA helix is shown by fast Fourier transformation in the upper right corner