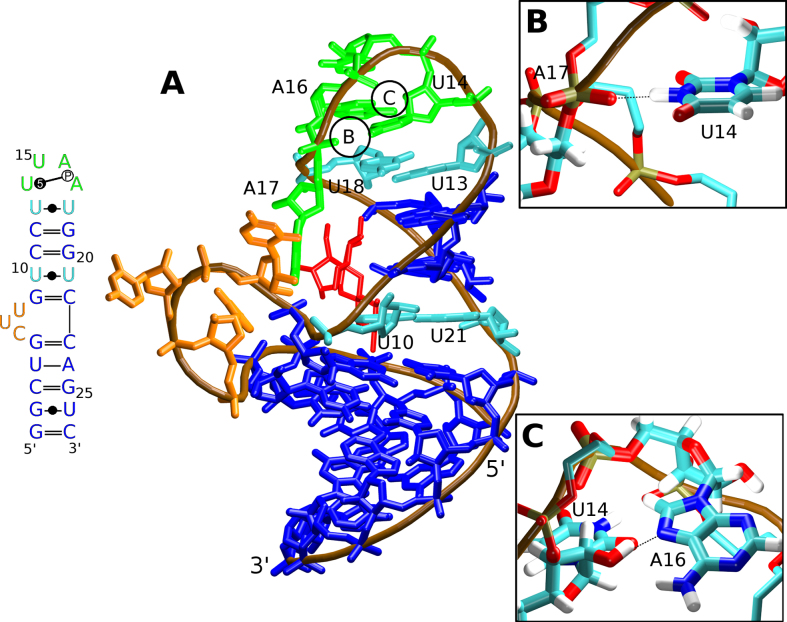
Figure 1.
(A) The secondary structure (left; standard Leontis & Westhof RNA base pair annotation extended by the classification of base-phosphate interactions is used, see also http://ndbserver.rutgers.edu/ndbmodule/ndb-help.html) (6,7) and the 3D representation (right) of the neomycin sensing riboswitch (PDB: 2n0j). The canonical and GU wobble base pairs are in blue, the cWW U/U base pairs are in cyan, the bulge is in orange, and the U-turn loop is in green. The RNA backbone is traced in brown and the bound ribostamycin ligand is shown in red. The chain termini and selected nucleotides are labeled. The circles B and C mark positions of the U14(N3)/A17(OP2) and U14(O2′)/A16(N7) signature H-bonds (the dashed black lines) with their structural details shown in the inset figures (B) and (C), respectively.