Fig. 1.
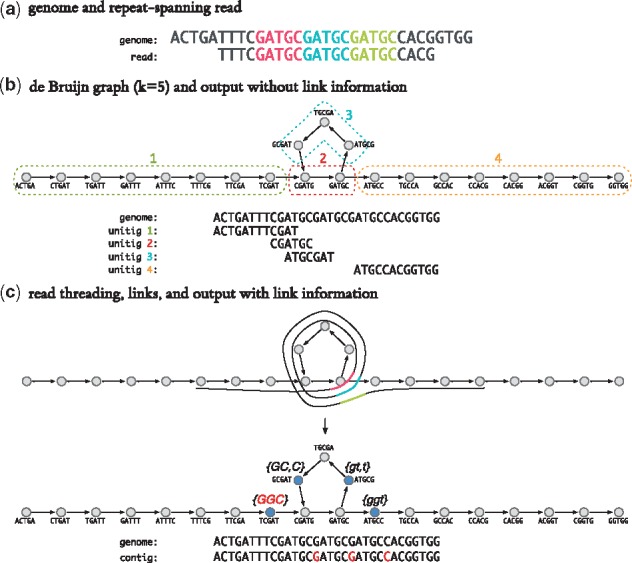
Utility of link information in traversing a graph cycle. (a) A 32-bp genome and a 23-bp read, each containing three (colour-coded) repeats of the 5-mer, GATGC. (b) The resulting de Bruijn graph (k = 5) with a repeat cycle, constructed from the genome sequence. The k-mers grouped by dashed boxes indicate the result of graph traversals to emit unitigs, with final sequences written below and positioned along the input genome for clarity. (c) Reads are ‘threaded’ (aligned) through the graph (top); the repeated k-mers are colour-coded. The alignment information is distilled to a set of junction choices to make when navigating the graph and stored as annotations on k-mers preceding junctions (middle). Multiple links are separated by a comma. Uppercase (lowercase) links indicate the choices to be made when traversing forwards (backwards). A k-mer’s links are picked up when we visit it. When we reach a junction, the next edge suggested by the oldest link(s) is taken, links that disagree are dropped, all remaining links trim off a junction choice and exhausted links are also dropped. The resultant contig recapitulating the entire genome is shown (bottom). Highlighted bases indicate the junction choices originating from the left-most link
