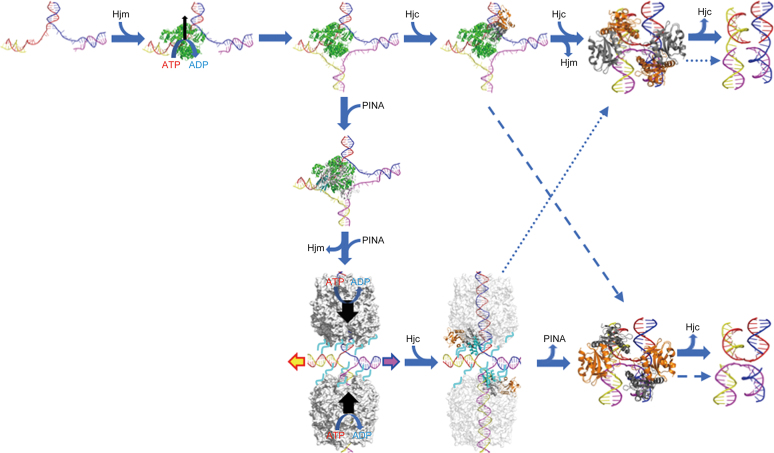
Figure 11.
A proposed model for PINA/Hjm/Hjc to process a stalled DNA replication fork. A stalled DNA replication fork (blue, red, yellow, and magenta strands) is recognized and bound by Hjm (green), which proceeds to regress the replication fork and form a HJ structure. Either Hjc (right) or PINA (down) is recruited to the HJ structure by Hjm. The Hjc dimer (gray and orange) binding to the HJ facilitates DNA handover from Hjm to Hjc, prompting Hjm dismissal and turning the open HJ into an X-HJ structure that is nicked by two Hjc dimers (PDB: 2WJ0). Alternatively, a PINA (white) monomer is recruited by Hjm through the interaction of Hjm and the PINA KH domain (cyan), leading to the assembly of the PINA hexamer on the HJ DNA and the dissociation of Hjm. Two PINA hexamers (white surface) are formed on two opposing HJ arms and mediate HJ migration powered by ATP hydrolysis. Two Hjc dimers then bind to the HJ and interact with the PINA C-terminal domains, destabilizing the PINA hexamers. After PINA hexamers are released, the two Hjc dimers turn the open HJ into the X-HJ structure and mediate two symmetric cuts. HJ migration directionality is indicated by colored arrows. Preferred HJ cleavages by Hjc are indicated with solid arrows while alternative HJ cleavages are indicated with dashed arrows. Unfolded PINA KH domains are indicated by cyan lines.