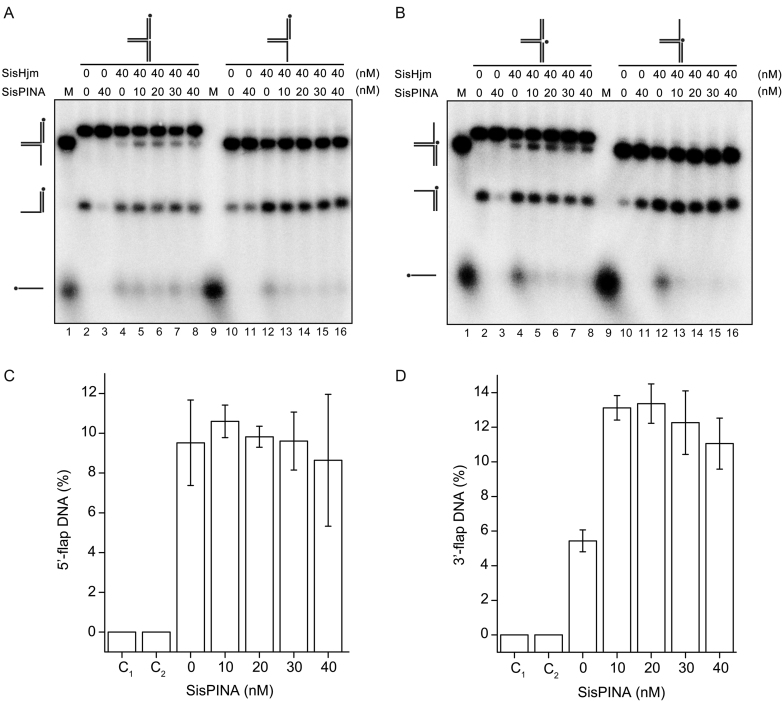
Figure 7.
SisPINA and SisHjm in combination remodel the replication fork. SisPINA and SisHjm were mixed to detect the capability of replication fork DNA processing. The fork substrates were labeled at the leading strand (A) or lagging strand (B) and the representative gels are shown. All the reactions were carried out at 45°C for 30 min. The ‘*’ indicates labeling of the substrates at the 5′ end. (C) Effect of SisPINA on the generation of 5′-flap DNA from a pseudo replication fork by Hjm. (D) Effect of SisPINA on the generation of 3′-flap DNA from a pseudo replication fork by Hjm. The products of 5′-flap DNA (A, lanes 2–8) and 3′-flap DNA (B, lanes 2–8) were quantified by Image J (NIH). The values were based on data of at least three experimental repeats.