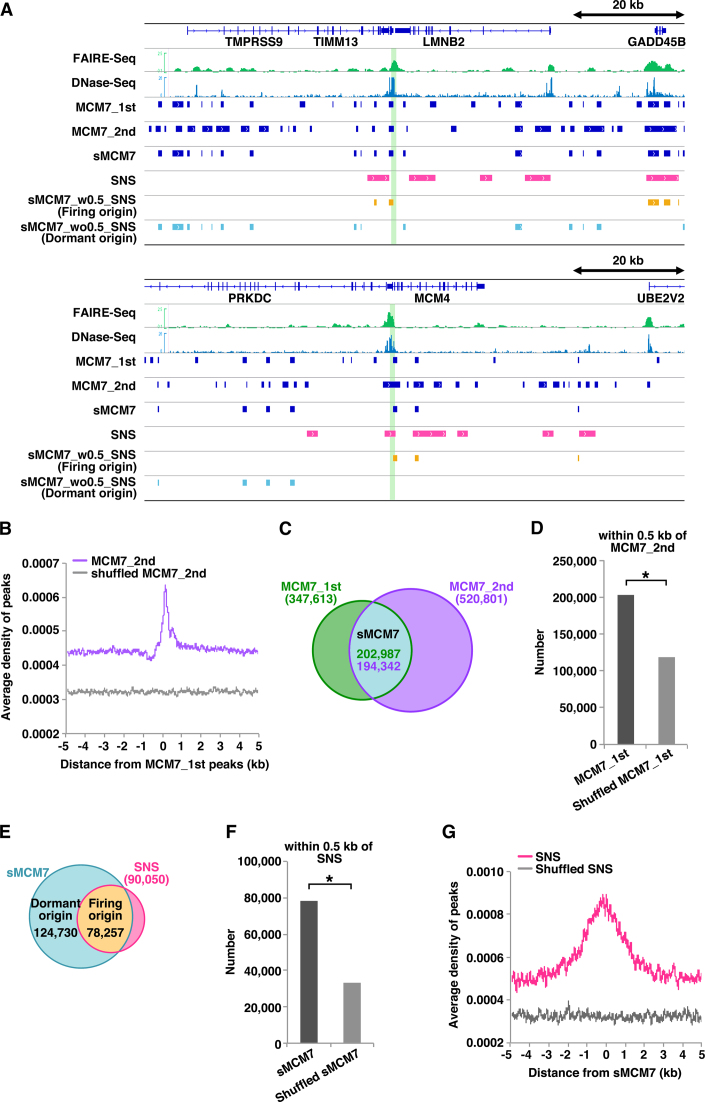
Figure 1.
Genome-wide identification of firing and dormant origin sites. (A) Selected snapshots of the genome browser view around the LMNB2 and MCM4 loci. Visual representations of FAIRE-Seq, DNase-Seq, MCM7 ChIP-Seq (first and second), SNS (13), sMCM7 (MCM7_1st_w0.5_MCM7_2nd), sMCM7_w0.5_SNS (firing origins) and sMCM7_wo0.5_SNS (dormant origins) data from HeLa cells are shown. Green lines indicate known origin regions. (B) Aggregation plots showing the localisation of MCM7_2nd ChIP-Seq peaks and shuffled peaks around MCM7_1st ChIP-Seq peaks. (C) Venn diagram showing the overlap (within 0.5 kb) of MCM7 peaks obtained in two independent experiments. (D) The number of MCM7_1st peaks within 0.5 kb of MCM7_2nd peaks is significantly higher than that obtained with shuffled MCM7_1st peaks. * indicates P < 0.0001 by Chi-square test. (E) Venn diagram showing overlap (within 0.5 kb) of sMCM7 and SNS peaks. (F) The number of sMCM7 peaks within 0.5 kb of SNS peaks is significantly higher than that obtained with shuffled sMCM7 peaks. * indicates P < 0.0001 by Chi-square test. (G) Aggregation plot showing the localization of SNS peaks and shuffled peaks surrounding sMCM7 peaks.