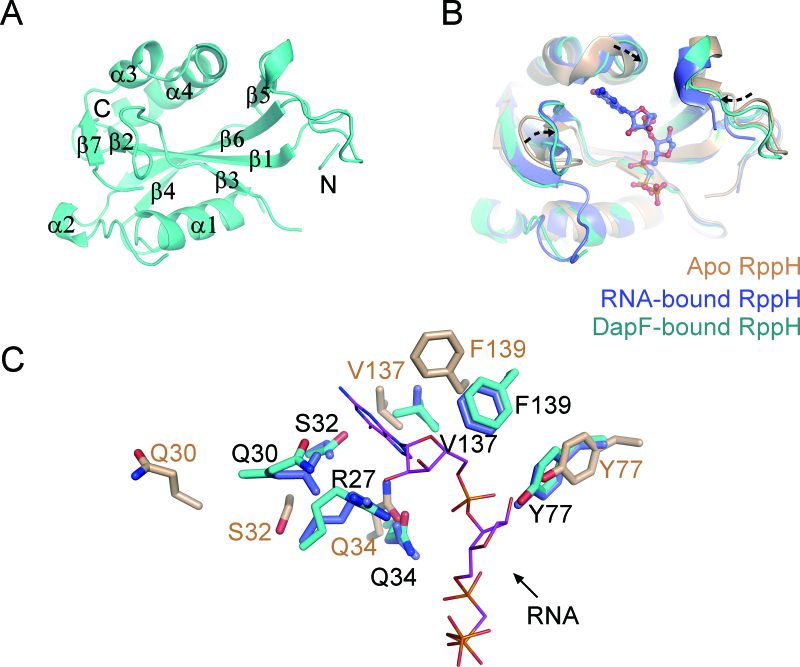
Figure 6.
The molecular basis of RppH activation by DapF. (A) A ribbon representation of the DapF-bound RppH (Mol A). The labels on the helices or strands are used to trace the RppH sequence. N-terminal and C-terminal ends are indicated for clarity. (B) Structural alignment of RppH in the apo state (PDB: 4S2V), RNA-bound state (PDB: 4S2Y) and DapF-bound state. Apo RppH is colored in brown, RNA-bound RppH in blue, and DapF-bound RppH in cyan. The diverse regions comprising a wobbling loop and two bent helices are indicated by dashed arrows, which are located in the vicinity of the pocket accommodating substrate RNA. (C) Side chain orientation of residues on the diverse region of RppH. Color representation of residues is the same as in diagram (B). RNA is shown as purple sticks. The residues that may be involved in RNA recognition in DapF-bound RppH nearly superimpose upon those of RNA-bound RppH and differ from those of apo RppH.