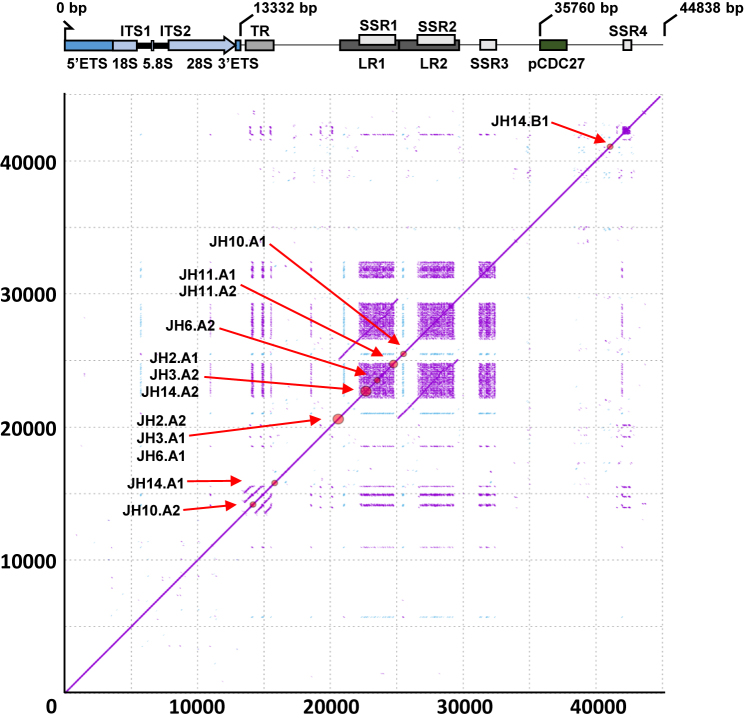
Figure 3.
Schematic diagram illustrating structure and sequence features of the new rDNA reference sequence. High-resolution (k = 15) self-similarity dot plot of the new reference sequence used for structural variant analysis. Purple dots indicate forward-strand matches, blue dots indicate reverse-complement matches. Red circles on the diagonal and labels show the positions of the identified structural variant breakpoints (clustered if distance ≤500 bp; see Supplementary Table S3 for precise coordinates and whole-genome validation status). Gene and sequence feature annotations are displayed below the x-axis. ETS: External transcribed spacer; 18S, 5.8 and 28S: core ribosomal DNA genes; ITS: Internal transcribed spacer; TR: Tandem repeats; LR1-2: Long repeats; SSR1-4: Simple sequence repeats; pCDC27: Pseudogene CDC27. Note that LR1 and LR2 are highly homologous, and that SSR1-4 are composed of near-identical sequence motifs.