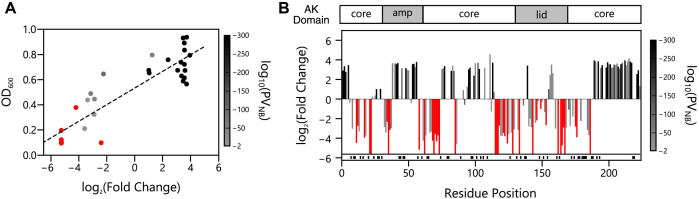
Figure 7.
Relationship between AK structure and retention of biological activity. (A) For thirty one variants, we compared the log2(fold change) values with growth complementation of Escherichia coli CV2 transformed with vectors that constitutively express each variant. This data displays a linear correlation (y = 0.066x + 0.533; R2 = 0.783). P-values obtained from the negative binomial model (PNB) are color coded and analyzed as described in Figure 6. (B) For each P variant, the log2(fold change) is shown as a function of the AK residue found at the N-terminus of the circularly permuted protein. The AK domain structure is shown as a frame of reference. Variants no longer observed in the selected library (infinitely diluted) are shown as bars that reach the line at the bottom of the graph. Red variants above the shaded region were observed in the selected library but were not significantly enriched (P-values > 0.01). Those cognate P and AP variant pairs absent from both the unselected and selected datasets (n = 52) are indicated as black lines shown below the x-axis.