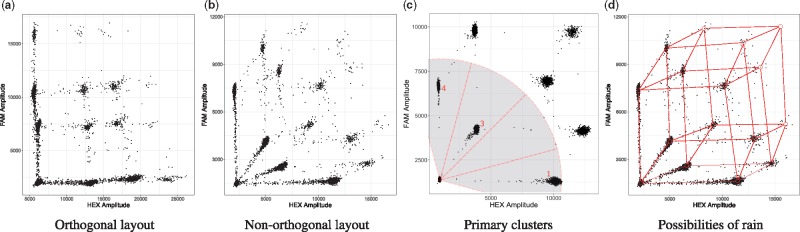
Fig. 1.
During a ddPCR run, each genetic target is fluorescently labelled with a combination of two fluorophores. The position of each droplet within this space reveals how many and, more importantly, which genetic targets it contains. Thus, droplets that contain the same targets, or the same combination of targets, cluster together. In clinical FFPE samples, DNA might be partially degraded, causing formation of rain and disappearance of the higher order clusters. (a) Multiplexing can cause overlap of clusters and rain. (b) Non-orthogonal layout avoids overlap of clusters and rain. (c) The angles between the droplets on the bottom left, which retain no target, and the primary clusters are highlighted. In case of genomic deletions or purposely missing clusters, it is possible to determine which cluster is missing. In this case, a genetic deletion of target 2 has occurred. (d) Graphical representation of the possible formation of rain along vectors