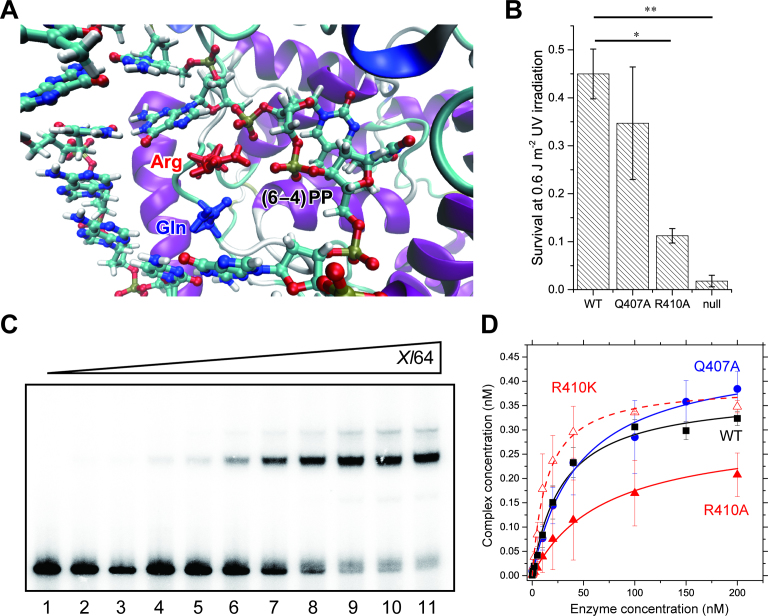
Figure 2.
Binding of the WT and mutants of Xl64 to the (6–4)PP containing substrate. (A) Selected view of the structure of Dm64 after the QM/MM optimization (see Materials and Methods). (B) UV survival assays of bacteria producing the WT or mutants of Xl64. Null means no production of Xl64 from the empty vector. The experiments were independently repeated three times (n = 3), and statistical analysis was carried out by the Student's t-test, where the significance was set to P < 0.01. Asterisks indicated the P-values of 0.0048 (*) and 0.0033 (**). (C) Electromobility shift assays of WT-Xl64. The upper bands represent the complex between 32P-DNA and the enzyme. Aliquots (0.5 nmol) of the 32P-labeled double-stranded 49-mer oligonucleotides were incubated with WT-Xl64 for 30 min at increasing concentrations of 0, 0.5, 1, 2, 5, 10, 20, 50, 100, 150 and 200 nM (lanes 1–11), and free- and bound-DNA were separated on a 5% polyacrylamide gel. The experiments were performed in triplicate, and the band intensities were quantified. (D) Binding of the WT (black square), Q407A (blue circle), R410A (red triangle), and R410K (open red triangle) of Xl64 to the (6–4)PP-containing double-stranded DNA. The complex concentrations were calculated from the quantified band intensities, and the mean values were plotted against enzyme concentrations. Their fitting curves are also shown.