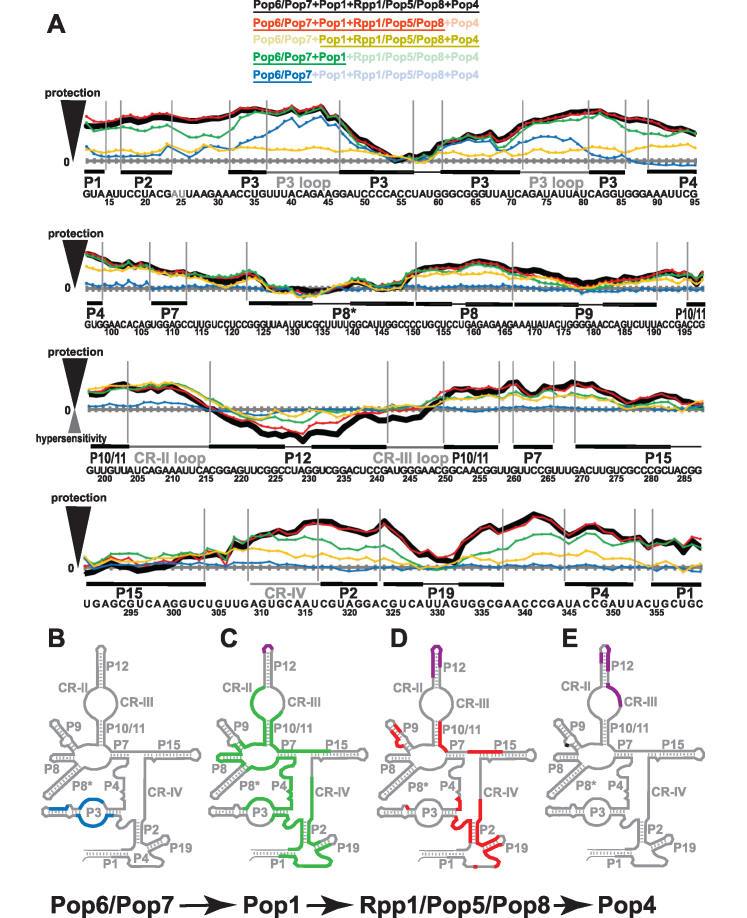
Figure 4.
Footprinting data for RNase P RNP assemblies. (A) Quantification of hydroxyl radicals (Fe(II)-EDTA) footprinting assays. The grey horizontal axes correspond to no additional protection (same reactivity as the RNA-only reference); the elevation above an axis reflects the degree of the protection (compared to RNA-only reference) in the presence of proteins. Protein compositions of the assayed RNP complexes are color-coded as shown on top of the figure; the proteins present in the RNP are underscored. Thick black line: proteins Pop6/Pop7, Pop1, Rpp1/Pop5/Pop8, Pop4. Red line: Pop6/Pop7, Pop1, Rpp1/Pop5/Pop8. Sand line: Pop1, Rpp1/Pop5/Pop8, Pop4. Green line: Pop6/Pop7, Pop1. Blue line: Pop6/Pop7. Sequence and secondary structure elements of the RNase P RNA (Figure 1A) are shown below the graphs. Helical stems are shown by thick solid lines; terminal loops are shown by thin solid lines; large internal loops and conserved elements are shown by dotted lines. See Supplementary Figure S8 for gels. (B–E) Changes in RNase P RNA reactivity induced by protein binding during RNase P RNP assembly. (B) Protection upon the binding of Pop6/Pop7 to RNA (blue). (C) Additional protection (green) and hypersensitivity (violet) upon the binding of Pop1 to the RNA-Pop6/Pop7 RNP. (D) Additional protection (red) and hypersensitivity (violet) upon the binding of Rpp1/Pop5/Pop8 to the RNA-Pop6/Pop7/Pop1 RNP. (E) Additional protection (black) and hypersensitivity (violet) upon the binding of Pop4 to the RNA-Pop6/Pop7/Pop1/Rpp1/Pop5/Pop8 RNP.