Figure 2.
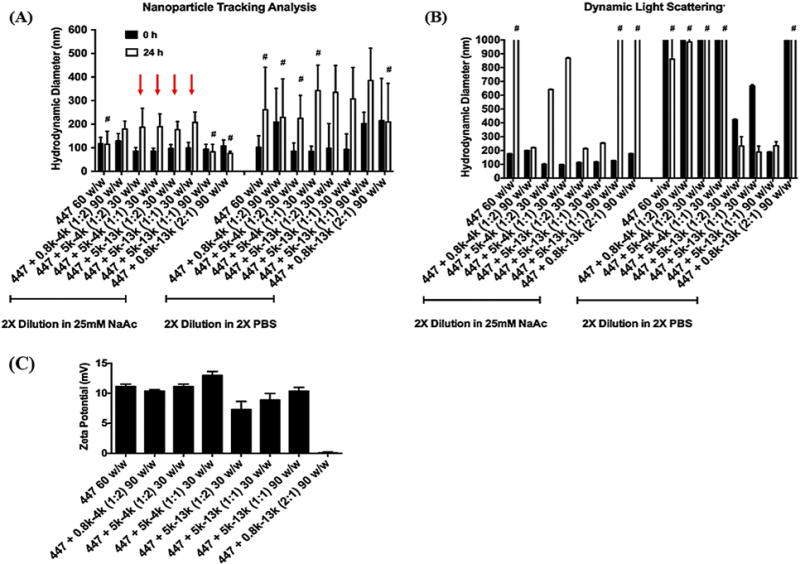
The size of polyplexes formed by self-assembly of enhanced green fluorescent protein (pEGFP) DNA with 447 alone or in combination with PEG-PBAE at various polymer:DNA and 447:PEG-PBAE w/w ratios was measured by (A) Nanosight (NTA) or (B) Zetasizer (DLS). The stability of the polyplexes was tested by sizing them after a 24-h incubation in either sodium acetate or PBS at room temperature. #: Indicates formulation conditions where polyplex aggregation is occurring, leading to unreliable size measurements (low particle concentration by NTA or greater than a micron in size by DLS). (C) The zeta potential of polyplexes. Data are mean ± SD of particle population for NTA and mean ± SD of 3 independent measurements for DLS. Red arrows indicate the four formulations that maintained particle stability and were selected for transfection evaluation.
