Fig. 2. Attenuated effects of tumor suppressor inactivation in Lkb1-deficient tumors further highlights a rugged fitness landscape.
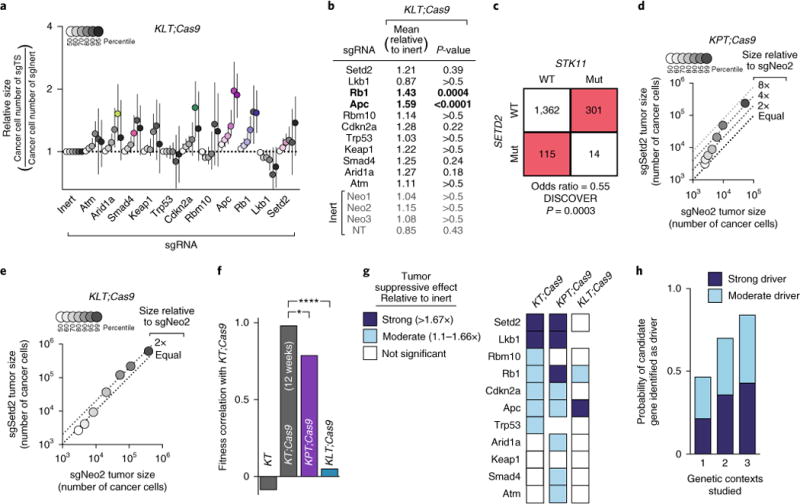
a, Tumor sizes at indicated percentiles for each sgRNA relative to the average of sgInert-containing tumors at the same percentiles. Merged data from 13 KT; Lkb1flox/flox; Cas9 (KLT;Cas9) mice 15 weeks after tumor initiation with Lenti-sgTS-Pool/Cre. Percentiles significantly different from sgInert are in color. Error bars denote 95% confidence intervals determined by bootstrap sampling. b, Estimates of mean tumor size, assuming a log-normal tumor size distribution, identified sgRNAs that significantly increase growth in KLT;Cas9 mice. Bonferroni-corrected, bootstrapped P-values are shown. P < 0.05 indicated by bold. c, Mutual exclusivity of STK11 (LKB1) and SETD2 mutations in human lung adenocarcinomas from TCGA and GENIE datasets (N = 1,792). WT, wild type; Mut, mutated. d, Tumor sizes in KPT;Cas9 mice with Lenti-sgSetd2/Cre-initiated tumors (N = 7) versus KPT;Cas9 mice with Lenti-sgNeo2/Cre-initiated tumors (N = 3). LN mean size is 2.4-fold greater in Lenti-sgSetd2/Cre-initiated tumors than Lenti-sgNeo2/Cre-initiated tumors and the 95th percentile of tumor size is 4.6-fold greater. e, Tumor sizes in KLT;Cas9 mice with Lenti-sgSetd2/Cre-initiated tumors (N = 7) versus KLT;Cas9 mice with Lenti-sgNeo2/Cre-initiated tumors (N = 5). Relative LN mean and relative 95th percentile are both significantly less than in Fig. 2d (P = 0.04 and P < 0.0001, respectively). f, Pearson correlations of fitness effect of tumor suppressors (determined by LN mean) across genetic backgrounds. sgTrp53 and sgLkb1 growth rates are excluded in KPT;Cas9 and KLT;Cas9 mice, respectively. *P = 0.04, ****P < 0.0001. g, Differential effect of each tumor suppressor gene with coincident Trp53 or Lkb1 deficiency. 95th percentiles that significantly deviate from sgInert tumors are in blue. h, Likelihood of identifying candidate tumor suppressors as drivers (as defined in g) versus the number of contexts studied. All possible genetic contexts were averaged.
