Fig.1. CRISPR indels.
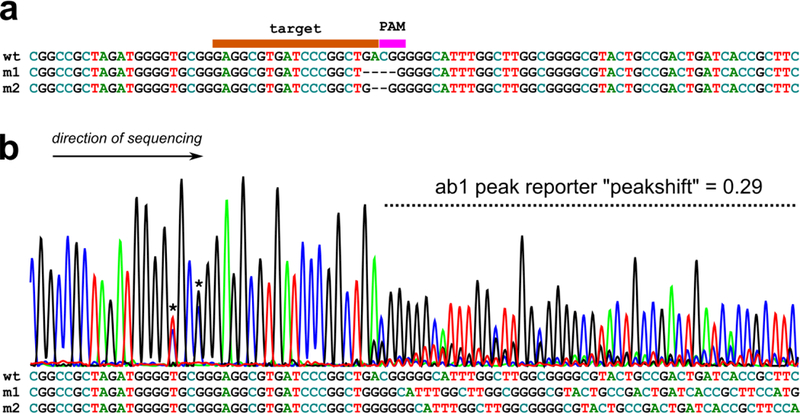
a) Wild-type (“wt”) target sequence aligned with two CRISPR knockout mutant sequences (“m1” and “m2”) generated by imprecise repair of CRISPR/Cas9-mediated double-stranded breaks. Alignment shows gaps (−) in place of missing nucleotides in target or PAM sequence. b) When sequenced by Sanger sequencing, pools of wild-type and mutant sequences will produce a “peakshift”, which can be quantified by ab1 Peak Reporter web app (see text for details). Below, the same sequences in (a) aligned without gaps, showing the cause of the overlapping peaks seen in the peakshift area. Asterisks denote naturally-occurring single-nucleotide polymorphisms.
