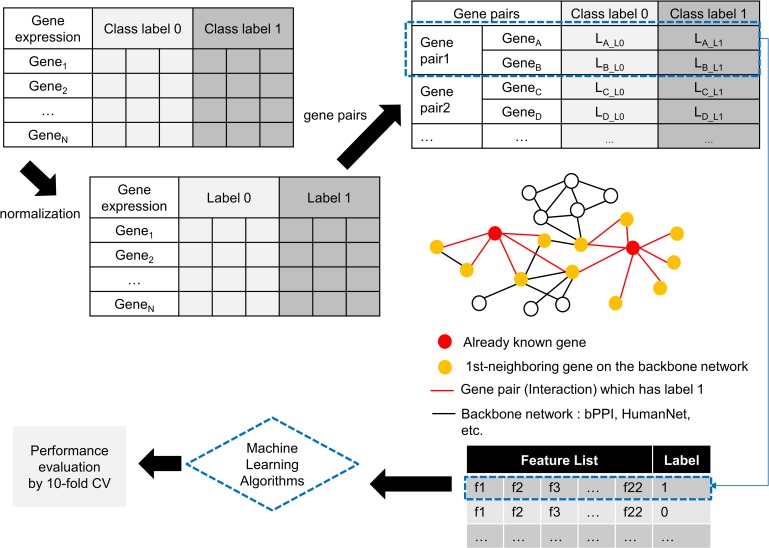
Fig 2. Overview of the proposed approach.
Gene expression data with two class labels are normalized by the z-scoring approach. For class label 1, which indicates disease, possible gene pairs are selected by incorporating disease-related genes and interactome data. For class label 0, which indicates normal, the same number of gene pairs as that for class label 1 is randomly selected. From all gene pairs, 22 features are extracted and used to inform the machine learning-based model. In order to evaluate performance, 10-fold cross validation is performed.