| Toxic compound |
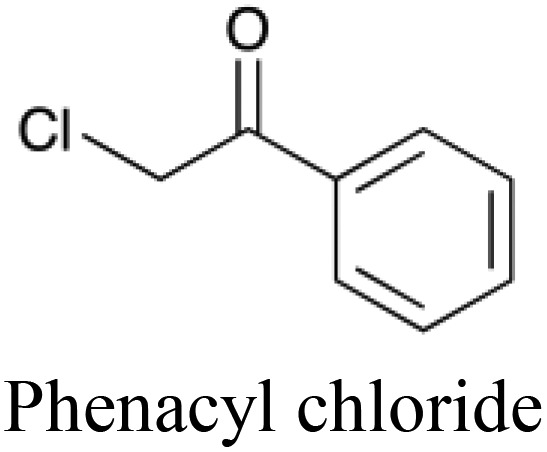
|
n/a |
| Non-toxic neighbors |
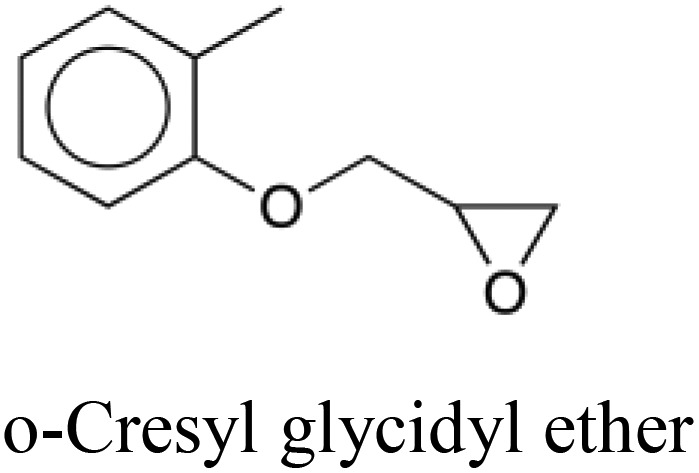
|
1. Protein kinase C zeta type |
| 2. Vesicular acetylcholine transporter
|
| 3. Macrophage metalloelastase
|
| 4. Bombesin receptor subtype 3 |
| 5. Heat shock protein HSP 90 beta |
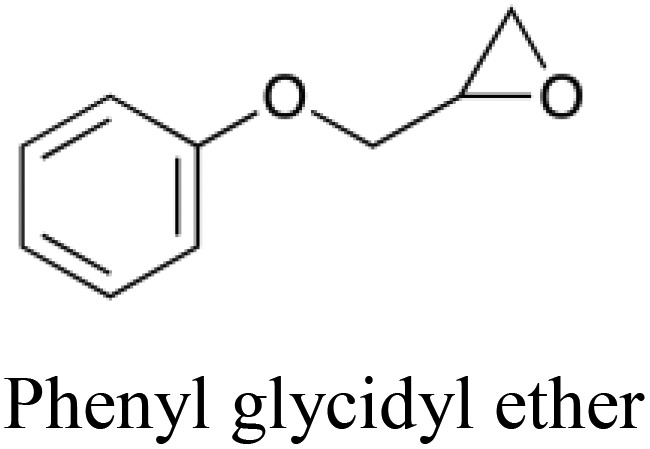
|
1. Vesicular acetylcholine transporter
|
| 2. Somatostatin receptor type 4 |
| 3. Protein kinase C zeta type |
| 4. Chymase |
| 5. Macrophage metalloelastase
|
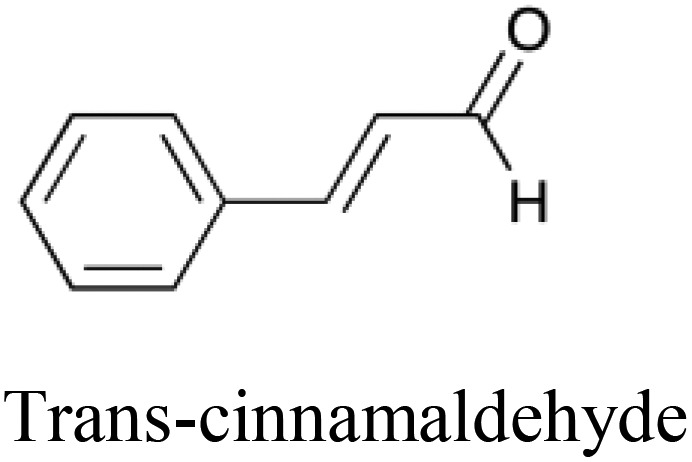
|
1. Vesicular acetylcholine transporter
|
| 2. Histamine H2receptor
|
| 3. Multidrug resistance protein 1 |
| 4. Macrophage metalloelastase
|
| 5. Sodium and chloride dependent glycine transporter 1 |
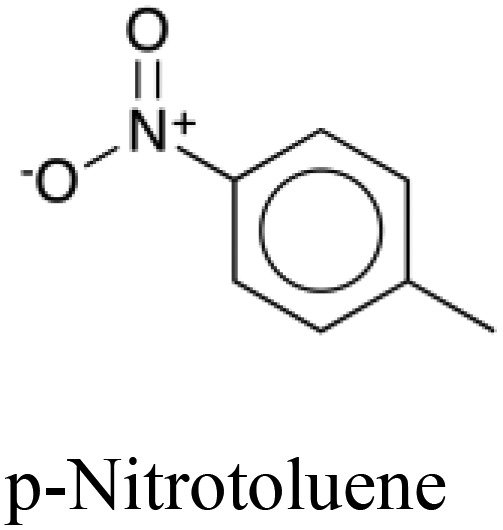
|
1. Histamine H2receptor
|
| 2. Bombesin receptor subtype 3 |
| 3. Heat shock protein HSP 90 beta |
| 4. Caspase 8 |
| 5. KiSS 1 receptor |
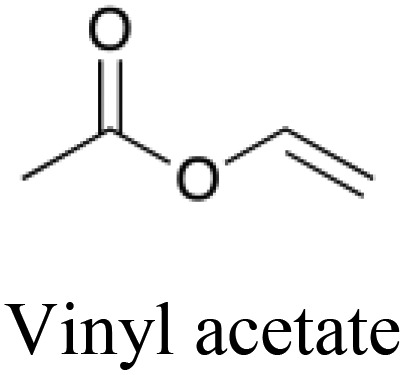
|
1. Histamine H2receptor
|
| 2. Vesicular acetylcholine transporter
|
| 3. Macrophage metalloelastase
|
| 4. Sodium and chloride dependent glycine transporter 1 |
| 5. Chymase |






